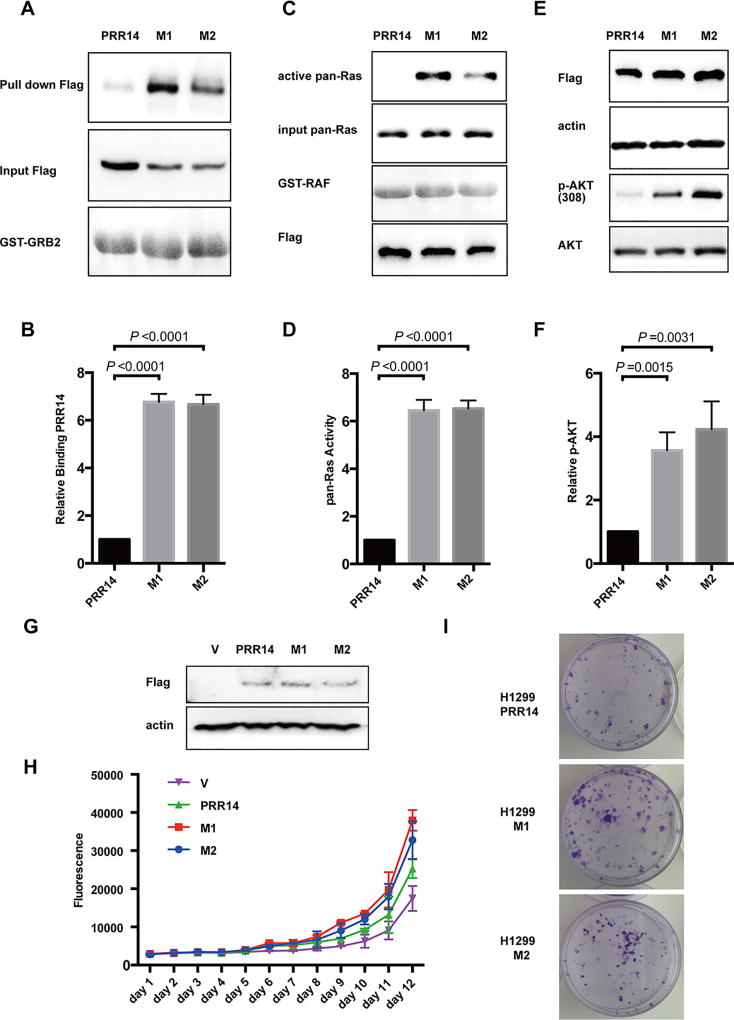
Fig 7. Mutants found in cancer patients increase interaction between PRR14 and GRB2.
(A) GST pull-down assay is employed to compare PRR14’s ability to interact with GRB2.The binding PRR14 protein was quantified and normalized to its total protein. The numbers of relative binding PRR14 protein are presented as the mean± S.D. (n = 3) and statistically analyzed by two-tailed student's t-test (B). (C) Ras pull-down activation Assay is used to compare PRR14’s ability to activate Ras in H1299 cells. The active Ras protein was quantified and normalized to total Ras protein. The numbers of relative abundance of active Ras protein are presented as the mean± S.D. (n = 3) and statistically analyzed by two-tailed student's t-test (D). (E) Mutants of PRR14 as well as their wild-type counterpart were transiently transfected into H1299 cells and cells were harvested for WB 24h later. The active Akt protein was quantified and normalized to its total protein, presented as the mean± S.D. (n = 3) and statistically analyzed by two-tailed student's t-test (F). (G) The overexpression of wild-type or mutant PRR14 is confirmed in H1299 PRR14, H1299 M1 and H1299 M2 cells lines by immunostaining. Growth curve (H) and colony formation assay (I) were also performed.