Figure 1.
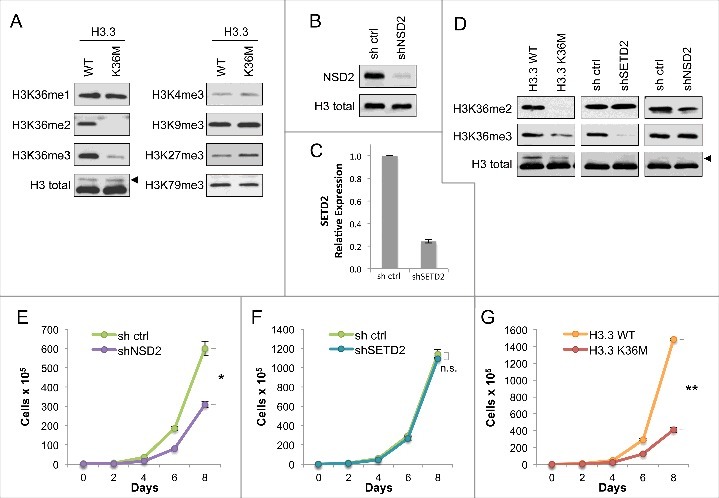
Expression of H3.3K36M depletes global di- and tri- methylation at H3K36 but recapitulates loss of H3K36me2 in cell proliferation (A) Western blot analysis lysates from HT1080 cells stably expressing H3.3 wild-type or K36M, using the indicated antibodies. Arrowhead, FLAG-H3.3 fusion construct appears as a higher molecular weight band than endogenous H3. (B) Western blot analysis of lysates from HT1080 cells stably expressing shNSD2 or control (sh ctrl) probed with anti-NSD2 or anti-H3 as a loading control. (C) Real-time PCR of SETD2 mRNA expression from RNA extracted from HT1080 cells stably expressing shSETD2 or sh ctrl. (D) Western blot analysis of H3K36 methylation levels in lysates from HT1080 cells stably expressing FLAG-H3.3 K36M mutant or KMT shRNA knockdown, using the indicated antibodies. Arrowhead, as in (A). (E-G) Proliferation assays of cells from (D), showing growth of cells expressing shNSD2 compared to control shRNA in (E), shSETD2 compared to control shRNA in (F), and H3.3K36M compared to H3.3WT control in (G). Cells were maintained in selection media and counted every two days for the duration of the assay. Error bars indicate s.e.m. from three experiments. p-values were calculated using a two-tailed Student's t test. *, p < 0.05. **, p < 0.01. n.s., not significant.
