Figure 1.
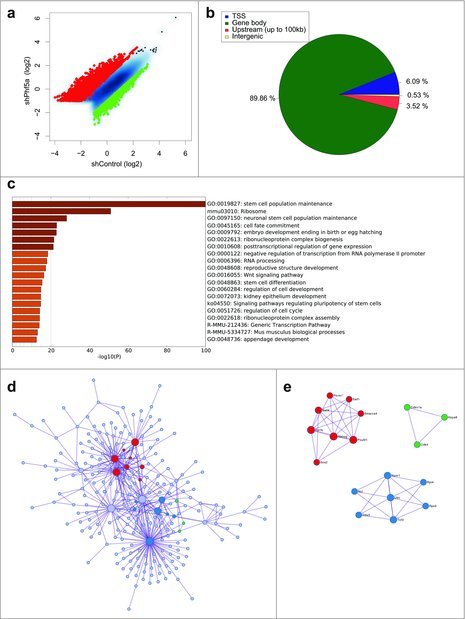
(a) Scatterplot of differentially regulated H2BK120-ub peaks in mouse ESCs following shControl or shPhf5a knockdown, respectively. Red: Transcripts that significantly gain H2BK120-ub densities. Green: Transcripts that significantly lose H2BK120-ub densities. (b) Binding profiles for genomic distribution of H2BK120-ub peaks (Transcription start site (TSS), Gene body, Upstream (up to 100kb) and intergenic) in ESCs, showing preferential (90%) binding within gene bodies. (c) Statistically enriched gene ontology terms and canonical pathways using Metascape (http://metascape.org) for all significantly altered H2BK120-ub transcripts after shPhf5a depletion. (d) Protein-protein interaction network of representative terms after hierarchical clustering analysis using MCODE.51 Each MCODE network node is assigned a unique color. Gene ontology enrichment analysis was applied to each MCODE network nodes to assign “meanings” to the network component. (e) Protein-protein interaction network of MCODE node components. Representative proteins include several core pluripotency factors that consist previously identified Paf1C targets.22
