Figure 1.
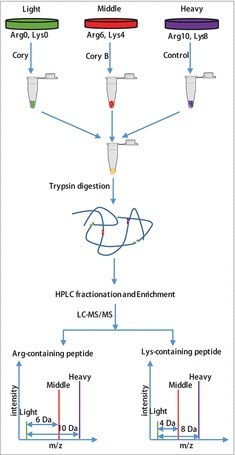
Flowchart of the identification of the quantitative phosphoproteome in neuronal autophagy. According to the weights of the labeled essential amino acids, N2a cells were divided into 3 groups, including “heavy” (13C615N2-Lysine, 13C615N4-Arginine), “middle” (13C414N2-Lysine, 13C614N4-Arginine) and “light” (12C614N2-Lysine, 12C614N4-Arginine). After checking the labeling efficiency (> 96%), the “light” and “middle” groups were treated with 15 μM Cory or Cory B for 3 h (Fig. S1), and the “heavy” group, which acted as a control group, was treated with 0.1% (v:v) DMSO. The cell lysates were mixed at a ratio of 1:1:1 and reduced with 10 mM dithiothreitol for 1 h at 56°C and then alkylated with 20 mM iodoacetamide for 45 min at room temperature and protected from light. Then, proteins were digested in solution with trypsin gold at a ratio of 1:50 (trypsin to protein) overnight and 1:100 (trypsin to protein) for another 4 h. After fractionation by high-pH reverse-phase HPLC, peptides were divided into 12 fractions. Finally, enriched phosphopeptides were subjected to LC-MS/MS for the identification of the quantitative phosphoproteome.
