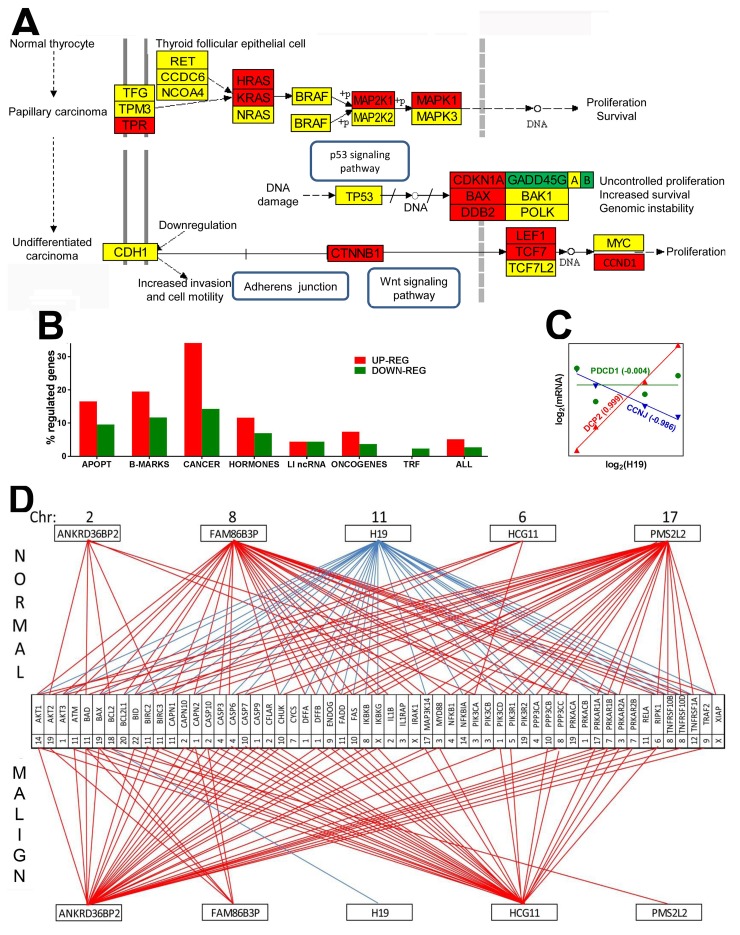
Figure 1. Papillary cancer of the thyroid regulates numerous genes and remodel transcriptomic networks.
(A) Regulation of the KEGG-determined pathway of thyroid cancer. Red/green/yellow background of the gene symbol indicates up-/down/no regulation. GADD45G|A|B indicates the three growth arrest and DNA-damage-inducible genes: gamma, alpha and beta. (B) Percentages of up- and down regulated genes in various groups. APOPT = apoptosis, B-MARKS = biomarkers, CANCER = thyroid cancer, HORMONES = thyroid hormones, LI ncRNAs = long intergenic non-protein coding RNAs, TRF = transcription factors, ALL = the entire transcriptome. (C) Examples of genes synergistically (DCPP2 = decapping mRNA 2), antagonistically (CCNJ = cyclin J) and independently (PDCD1 = programmed cell death 1) expressed with H19 (= H19, imprinted maternally expressed transcript, long non-coding RNA). Numbers near gene symbols are Pearson pair-wise correlation coefficients between the (log2) expression levels of H19 and linked gene. (D) Example of remodeling of the transcriptomic networks by which ncRNAs regulate apoptosis. Red/blue line indicates significant (p-val < 0.05) synergism/antagonism between the linked gene and ncRNA. Numbers close to genes and ncRNAs’ symbols are their hosting chromosomes (Chr).