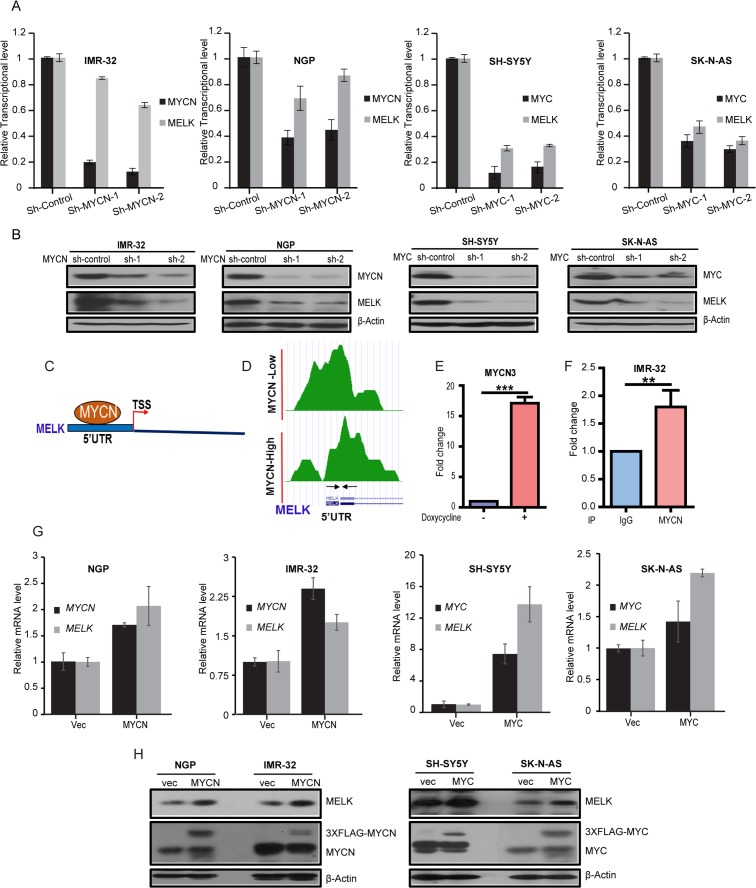
Figure 2. MELK is a transcription target of MYCN/MYC in NB.
(A) Stable sh-RNA-mediated knockdown of MYCN/MYC. The qPCR analyses of MYCN/MYC and MELK expression levels were shown. (B) Total proteins of MYCN/MYC and MELK were measured in the indicated cells. (C and D) MYCN ChIP-seq analysis showed binding peaks of MYCN on MELK 5’UTR. TSS- Translation Start Site. Designed primers for ChIP-seq analysis is shown with arrows under the peaks. These primers encompass the E-Box motif. (E) ChIP-qPCR analysis for MYCN binds on MELK 5’UTR site in MYCN3 cell line. Fold change with or without doxycycline treatment on MYCN3 cell line was shown. (F) ChIP-qPCR analysis for MYCN binds on MELK 5’UTR site in IMR-32 NB cell line. IP-IgG and IP-MYCN of MELK-ChIP assay on IMR-32 cells were shown. (G) qPCR analysis of MYCN/MYC and MELK mRNA levels in NB cell lines with stable MYCN/MYC-overexpression were shown. (H) MYCN/MYC and MELK protein levels in NB cell lines with stable MYCN/MYC-overexpression were shown.