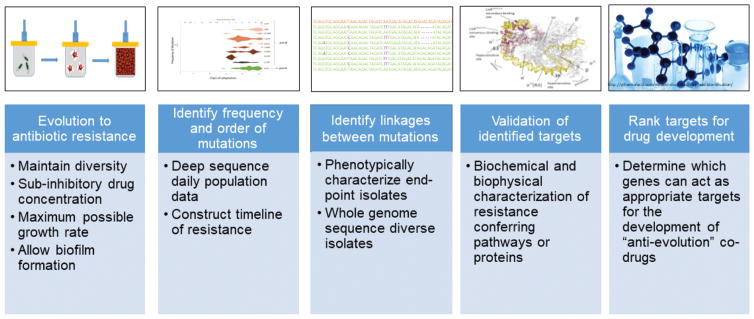
Figure 2. Pipeline of quantitative experimental evolution to predict antibiotic resistance and identify targets for drug discovery.
Each stage produces essential data and approaches that when taken together predict resistance, identify the most important targets, suggests potential biochemical mechanisms, and leads to assay development. The stages are: 1) Evolve resistant populations in a bioreactor under polymorphic selection conditions; 2) Identify the frequency and order of mutations correlated with antibiotic resistance as a function of time; 3) Identify the genotypes of end-point isolates to establish genetic linkages; 4) Validate the effect of mutations by physicochemical characterization; 5) Rank candidates for potential drug development.