Abstract
Objective
Reactive arthritis (ReA) is an inflammatory disorder occurring several weeks after gastrointestinal or genitourinary infections. HLA-B27 positivity is considered a risk factor, though not necessarily predictive of disease incidence. Among non-genetic factors, the intestinal microbiome may play a role in disease susceptibility. The objective of this study was to characterize the gut microbiota and host gene interactions in ReA and post-infectious spondyloarthritis.
Methods
Adult peripheral spondyloarthritis and control subjects with preceding infections that did not develop arthritis were prospectively recruited from a highly prevalent geographic region. Clinical variables, HLA status, and 16S rRNA gene sequencing of intestinal microbiota were analyzed.
Results
ReA subjects showed no significant differences from controls in gut bacterial richness or diversity. However, there was a significantly higher abundance of Erwinia and Pseudomonas, and increased prevalence of typical enteropathogens associated with ReA. Subjects with ultrasound evidence of enthesitis were enriched in Campylobacter, while subjects with uveitis and radiographic sacroiliitis were respectively enriched in Erwinia and unclassified Ruminococcaceae, and both enriched in Dialister. Host genetics, particularly HLA-A24, were associated with differences in gut microbiota diversity irrespective of disease status. We determined several co-occurring taxa that were also predictive of HLA-A24 status.
Conclusion
This is the first culture-independent study characterizing the gut microbial community of post-infectious arthritis. Although bacterial factors correlated with disease presence and clinical features of ReA, host genetics also appeared to be a major independent driver of intestinal community composition. Understanding of these gut microbiota host-genetic relationships may further clarify the pathogenesis of post-infectious spondyloarthropathies.
Keywords: reactive arthritis, spondyloarthritis, microbiome
INTRODUCTION
Reactive arthritis (ReA) is an inflammatory disorder that manifests in young adults, usually a few weeks after a gastrointestinal (GI) or genitourinary (GU) tract infection. While it is typically self-limited and resolves within months, a proportion of individuals develop persistent symptoms. ReA is classified as one of several spondyloarthritidies (SpA). Symptoms include asymmetric oligoarthritis (most often of the large joints); extra-articular features, including enthesitis, tendinitis, uveitis; and a variety of skin rashes such as erythema nodosum, keratoderma blenorrhagica and circinate balanitis (1). The classic enteropathogenic bacteria associated with ReA are Yersinia, Salmonella, Shigella, and Campylobacter, while Chlamydia is the most common etiology following GU infections (1). However, Escherichia coli (2) and Clostridium difficile (3) have also been implicated.
The mechanism for post-infectious peripheral SpA and ReA development remains elusive. HLA-B27 positivity has long been studied as a genetic risk factor for the disease (4). It is highly prevalent in Caucasian Northern Europeans, but virtually absent in other populations that develop ReA (5). In fact, several recent epidemiologic studies estimate that no more than half of individuals who develop ReA are HLA-B27 positive (6), while the presence of the allele has poor predictive value for the development of disease (7). Furthermore, cases of ReA are seldom diagnosed in the United States (8), and the ones encountered are typically in immigrant populations (9). However, ReA is much more prevalent in the native population of Central America, and particularly in Guatemala, where HLA-B27 positivity is low (10).
Unlike septic arthritis, which directly results from joint infection by pathogenic microorganisms, synovial fluid cultures in ReA are sterile, implying the possibility of an autoimmune phenomenon. Several models for ReA pathogenesis have been proposed. They include the persistence of causative agents that trigger an exaggerated immune response; intracellular uptake and trafficking of bacteria (or their components) into the synovium causing inflammation; biomolecular mimicry, whereby bacterial antigens have host immunologic homology but are different enough to induce an immune response in synovial fluid; Toll-like receptors (TLR), particularly TLR-2, recognizing bacterial ligands and activating an immune cell response; and intestinal infection resulting in gut permeability, allowing luminal antigens to interact with the host immune system and cause distal joint inflammation (11, 12). In support of the latter, recent investigations indicate that synovial fluid of individuals with ReA contains potentially immunogenic products such as bacterial DNA, antigenic proteins, and lipopolysaccharides (13, 14).
Another potential risk factor that has not been explored is the collection of commensals and pathogenic microorganisms residing in the gut, namely the intestinal microbiome. Given the close relationship between gut infection and ReA, the microbiome likely plays a role in susceptibility to disease. To date, there have not been any studies looking at the intestinal microbiota as it relates to ReA or post-infectious peripheral SpA development. The objective of this study was to characterize the gut microbiota of subjects who had previous GI or GU infections in a highly prevalent area, and evaluate for differences between those who developed post-infectious peripheral SpA and those who did not. We also explored whether clinical phenotype, host genetics, and treatment strategies were associated with significant intestinal microbiota perturbations.
MATERIALS AND METHODS
Study participants
Thirty-two ReA (post-infectious peripheral SpA) and thirty-two control subjects were prospectively enrolled from July to October 2014 in Guatemala City, Guatemala (a highly prevalent area for ReA) through the Guatemala-Penn Partners Initiative (15). ReA subjects were recruited from two rheumatology clinics—AGAR and CMER. Controls were recruited from primary care clinics. Inclusion criteria were age 18–55 and preceding GI, GU, or sexually transmitted infection within 3–6 months prior to enrollment. All preceding infections and antibiotic use were subject-reported. Subjects with GI infections had preceding diarrhea, and those with GU infections had dysuria, frequency, and occasionally hematuria. Stool or urine culture at the time of infection was not available. Exclusion criteria were an alternative diagnosis of inflammatory bowel disease (IBD), psoriasis, another inflammatory arthritis or autoimmune disease, and/or active malignancy. ReA was diagnosed by a study rheumatologist and in accordance with the Assessment of Spondyloarthritis International Society (ASAS) criteria for peripheral SpA (16), based on the triad of characteristic musculoskeletal findings, patient reported history of GI/GU infections and lack of evidence for alternative diagnosis. Although we use the term ReA throughout, our patient population can be more accurately defined under the broader category of post-infectious peripheral SpA, given the absence confirmatory cultures/serology.
Subject evaluation and sample collection
After obtaining informed consent, all subjects were evaluated by a rheumatologist who took a detailed clinical history and performed a physical and musculoskeletal exam. History of uveitis was subject-reported, though 75% of cases were diagnosed by an ophthalmologist and subsequently referred to rheumatology clinic. Blood was obtained for standard laboratory testing and HLA-typing, while fecal samples were collected for microbiota analysis around the time of study enrollment. Cultures of specific bacteria were not performed. All subjects also had a plain radiograph of the sacroiliac joints and an ultrasound of the bilateral Achilles tendons. Further details are described in the Supplementary Materials.
DNA extraction and sequencing
Fecal samples were stored at −80°C until processing. Bacterial DNA was extracted using the MoBio Power Soil kit (MoBio) and bead-beater for mechanical disruption following a previously described protocol (17). To determine gut microbiota composition, high-throughput DNA sequencing of the V4 hypervariable region of bacterial 16S rRNA was performed using the MiSeq Illumina platform (150 bp read length, paired-end protocol) at the NYU Genome Technology Center. Further details are described in the Supplementary Materials.
Sequence analysis
The 16S rRNA sequences were analyzed using the Quantitative Insights into Microbial Ecology (QIIME) pipeline v1.9.1 (18) and R v3.3.1 (19). Reads were demultiplexed and sequences clustered into operational taxonomic units (OTUs) using closed-reference OTU picking based on ≥97% similarity with the Greengenes reference database (20). Each OTU count was normalized to the total sum of counts in the sample. QIIME and the vegan library in R were used to construct alpha diversity plots to measure the diversity within a sample by looking at species richness (number of species present) and evenness (relative abundance of different species). Weighted UniFrac was used to construct beta diversity of bacterial communities to measure the distance or dissimilarity between sample pairs (21). Beta diversity is represented by principal coordinate analysis (PCoA). Further details are described in the Supplementary Materials.
LEfSe
Bacterial taxa whose relative abundances were significantly different between groups were identified with Linear discriminant analysis Effect Size (LEfSe) (22). A log LDA score >2 was the threshold for discriminating between phenotypic groups. Given that LEfSe does not use multiple hypothesis testing, False Discovery Rate (FDR) analysis with a threshold value of <0.2 and Bonferroni analysis with a threshold value of <0.05 were subsequently applied to identify the main differentiating taxa.
Co-occurrence network analysis
OTUs with more than one count were selected and clustered at the family level. Correlations between families were calculated using SparCC (23) with 20 iterations and 500 bootstraps. Non-significant correlations (p<0.05, two-sided) were then discarded. The remaining correlations were loaded into Cytoscape v3.0.2 (24) to be visualized as a co-occurrence network, where each node represents a bacterial family and edges between nodes represents their correlations, with shorter edges indicating stronger correlations. The network was displayed using the edge-weighted spring embedded metrics layout, with positive correlations colored blue and negative ones in gray. We then searched for maximal cliques using the Bron-Kerbosch algorithm (25) as implemented in the NetworkX python package (26) to identify subsets of bacteria that belong to the same clique, which may be reflective of synergistic cooperation among the taxa.
Statistical analysis
Statistical analyses were performed with R and GraphPad Prism v7.0b (27). For baseline characteristics, chi-square was used for dichotomous variables and the independent t-test for continuous variables. Chi-square was also used to calculate differences in prevalence of usual intestinal enteropathogens between ReA and control subjects. The Mann-Whitney U test was used to calculate differences in alpha diversity and relative abundance of specific taxa that were grouped by disease/HLA status when comparisons were made between two variables. The Kruskal-Wallis test was used when comparisons were made among three variables. The Adonis (Permanova) test was used to calculate differences in beta diversity. P-values <0.05 were considered significant.
IRB approval
This study was approved by the Institutional Review Board of the University of Pennsylvania (#819438) and the ethics committee of Universidad Francisco Marroquin. Further details are described in the Supplementary Materials.
RESULTS
Characteristics of the study population
Demographics of the study population are shown in Table 1. A total of 64 subjects were prospectively enrolled in the study; 32 had post-infectious peripheral SpA and 32 served as controls. All the subjects reported preceding GI and/or GU infections 3–6 months prior to enrollment. None reported sexually transmitted infections. Treatment of these infections with antibiotics was reported in 62.5% of cases and 46.9% of controls. The majority of subjects were female and racially identified as Mixed (Mestiza) or Native (Indigena). Very few participants carried an HLA-B27 allele.
Table 1.
Clinical and laboratory characteristics of the study population. To calculate statistical significance, chi-square was used for dichotomous variables and the independent t-test was used for continuous variables.
| ReA† (n = 32) |
Control (n = 32) |
p-value | |
|---|---|---|---|
|
| |||
| Age (years) | 0.268 | ||
| Range | 18–55 | 18–54 | |
| Mean ± SD | 39 ± 11.5 | 36 ± 9.9 | |
|
| |||
| Gender (%) | 0.095 | ||
| Male | 6 (18.8%) | 12 (37.5%) | |
| Female | 26 (81.3%) | 20 (62.5%) | |
|
| |||
| Race (%) | 0.050 | ||
| Caucasian (Blanca) | 1 (3.1%) | 7 (21.9%) | |
| Native (Indigena) | 15 (46.9%) | 9 (28.1%) | |
| Mixed (Mestiza) | 16 (50.0%) | 16 (50.0%) | |
|
| |||
| BMI | 0.423 | ||
| Range | 19.0 – 34.6 | 17.6 – 36.1 | |
| Mean ± SD | 25.1 ± 4.4 | 26.0 ± 4.3 | |
|
| |||
| Disease characteristics | |||
| Peripheral arthritis | 32 (100.0%) | 0 (0.0%) | <0.0001 |
| Ultrasound enthesitis | 14 (43.8%) | 2 (6.3%) | 0.0005 |
| Radiographic sacroiliitis | 18 (56.3%) | 16 (50.0%) | 0.616 |
| Uveitis | 20 (62.5%) | 2 (6.3%) | <0.0001 |
|
| |||
| HLA-typing (%) | |||
| A2 positive | 14 (43.8%) | 16 (50.0%) | 0.616 |
| A24 positive | 14 (43.8%) | 11 (34.4%) | 0.442 |
| B15 positive | 2 (6.3%) | 2 (6.3%) | * |
| B27 positive | 2 (6.3%) | 2 (6.3%) | * |
| B35 positive | 19 (59.4%) | 15 (46.9%) | 0.316 |
| C7 positive | 17 (53.1%) | 15 (46.9%) | 0.617 |
|
| |||
| Infection (%) | |||
| Gastrointestinal | 23 (71.9%) | 29 (90.6%) | 0.055 |
| Genitourinary | 10 (31.3%) | 4 (12.5%) | 0.070 |
|
| |||
| Antibiotics (%) | 20 (62.5%) | 15 (46.9%) | 0.209 |
|
| |||
| Sulfasalazine (%) | 3 (9.4%) | 0 (0%) | * |
ReA based on ASAS criteria of post-infectious peripheral SpA; no confirmatory cultures or serology available for definite ReA;
Not enough power to detect a statistical difference.
All ReA subjects had peripheral arthritis (100% ReA, 0% controls; p<0.0001), with an average tender joint count of 5 (predominantly large joints). Ultrasound-identified Achilles enthesitis was seen in 14 ReA subjects (43.8% ReA, 6.3% controls p=0.0005). The average number of tender entheses was 5, with the Achilles tendon being the most common. Radiographic sacroiliitis was demonstrated in 18 ReA subjects (56.3% ReA, 50.0% controls; p=ns). These were mostly grades I and II, showing sclerosis without presence of erosions. Finally, 20 subjects reported a prior diagnosis of uveitis (62.5% ReA, 6.3% control; p<0.0001). Additional demographic data is described by Garcia Ferrer, et al. (28) and currently under review for publication.
Gut microbiota diversity in ReA and control subjects
A total of 64 fecal samples were obtained and sequenced from ReA and control subjects. The mean number of reads per sample after sequencing was 74,382 (range 46,483 – 122,511, median 71,233). Utilizing a distance-based similarity of ≥97% to define OTUs, we identified a total of 5,356 unique OTUs. There were no statistically significant differences in alpha diversity (within subject diversity as estimated by the total number of unique OTUs, the Shannon diversity index, and the Faith’s phylodiversity index) between ReA and control subjects (Supplementary Figure 1A–C). However, there was a trend toward overall decreased diversity in the ReA group using the Shannon index (p=0.069), a finding shared with psoriatic arthritis (PsA) (29). Evaluation of microbial similarity between ReA and control subjects using PCoA based on weighted UniFrac distances showed no distinctive clustering patterns among samples (Supplementary Figure 1D). Furthermore, there were no statistically significant differences in alpha or beta diversity when subjects were grouped by whether they had received antibiotics to treat their preceding infection (Supplementary Figure 2A). There was not enough power to detect a difference in the case of sulfasalazine (Supplementary Figure 2B), a frequently used medication with known antibiotic properties.
Specific intestinal taxa differentiate ReA samples and correlate with phenotypic manifestations
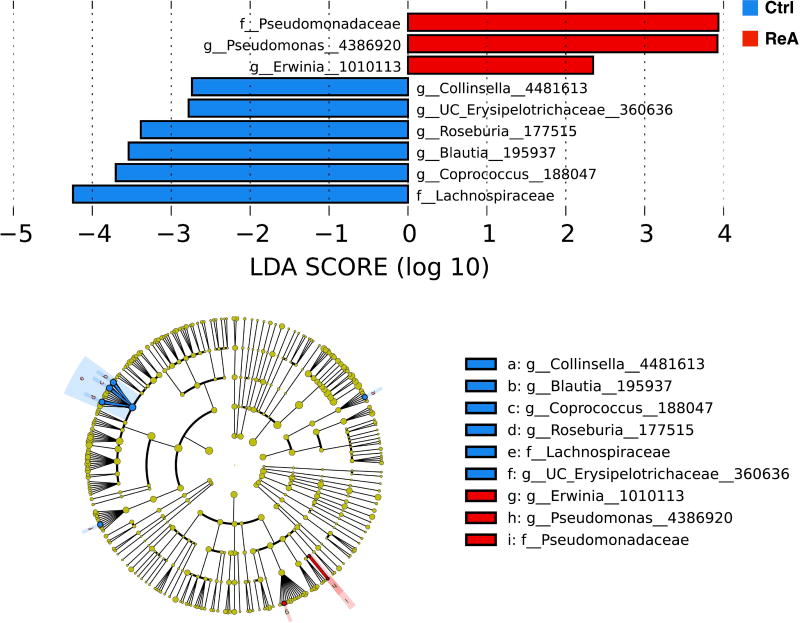
We applied LEfSe analysis to determine whether specific individual bacterial taxa were differentially enriched in ReA when compared to controls (Figure 1). ReA subjects had a statistically significant higher abundance of Erwinia and Pseudomonas, two known intestinal enteropathogens. This finding contrasted with control individuals, where the relative abundance of several genera, most of them considered to be commensals, were enriched, including Blautia, Coprococcus, Roseburia, and Collinsella (log LDA>2; none passed FDR or Bonferroni correction thresholds).
Figure 1.
LEfSe analysis of reactive arthritis (ReA) and control (Ctrl) subjects. LDA demonstrates that enteropathogens were enriched in ReA subjects, while commensals were enriched in control subjects. Cladogram shows the relationships among differentiating taxa between subject groups. Log LDA >2 for all taxa. None passed FDR or Bonferroni correction thresholds.
To further characterize the presence of intestinal enteropathogens typically associated with ReA, we focused our attention on sequences belonging to Salmonella, Shigella, Campylobacter, and Erwinia (a taxon with ≥97% identity to various species of Salmonella, Shigella, and Yersinia by BLAST search). Interestingly, while less than half of control subjects carried any of these taxa, 71.9% of subjects with disease had sequencing evidence of at least one such bacteria (p=0.042; Supplementary Figure 3).
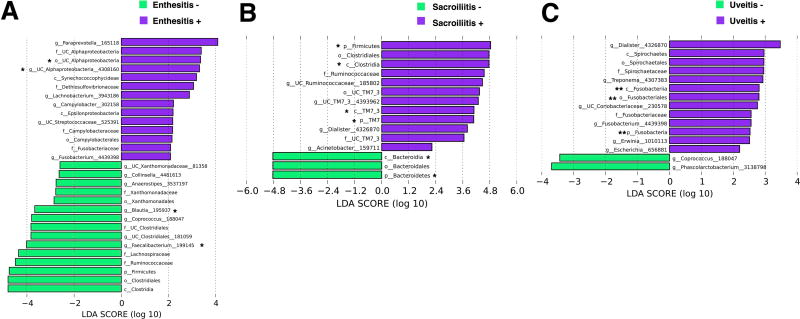
Clinical and radiographic metadata showed that alpha diversity was significantly lower in subjects with ultrasound evidence of enthesitis (Supplementary Figure 4A) and beta diversity was significantly different in subjects with radiographic evidence of sacroiliitis (Supplementary Figure 4B). There were no diversity differences in subjects with or without uveitis (Supplementary Figure 4C). However, specific taxa were enriched when these manifestations were present, as shown by LEfSe analysis. In particular, unclassified Alphaproteobacteria (genus and order) was enriched in subjects with enthesitis (Figure 2A), while Clostridia (class), TM7_3 (class), Firmicutes (phylum), and TM7 (phylum) were enriched in subjects with sacroiliitis (Figure 2B), all passing FDR correction threshold. Fusobacteriales (order), Fusobacteria (class), and Fusobacteria (phylum) were enriched in subjects with uveitis, passing both FDR and Bonferroni correction thresholds (Figure 2C). Although not achieving FDR or Bonferroni significance, Campylobacter was overrepresented in subjects with enthesitis (Figure 2A), while unclassified Ruminococcaceae was overrepresented in subjects with sacroiliitis (Figure 2B) and Erwinia in subjects with uveitis (Figure 2C). Interestingly, the genus Dialister was independently enriched in subjects with sacroiliitis (Figure 2B) and those with uveitis (Figure 2C).
Figure 2.
LEfSe analysis for clinical and radiographic data specific to reactive arthritis. Unclassified Alphaproteobacteria (genus and order) were enriched in subjects with ultrasound evidence of enthesitis (A), while Clostridia (class), TM7_3 (class), Firmicutes (phylum), and TM7 (phylum) were enriched in subjects with radiographic sacroiliitis (B), all passing FDR correction threshold. Fusobacteriales (order), Fusobacteria (class), and Fusobacteria (phylum) were enriched in subjects with uveitis, passing both FDR and Bonferroni correction thresholds (C). Although not achieving FDR or Bonferroni significance, Campylobacter was enriched in subjects with enthesitis (A), unclassified Ruminococcaceae was enriched in subjects with sacroiliitis (B), and Erwinia was enriched in subjects with uveitis (C). Dialister was independently enriched in subjects with sacroiliitis (B) and those with uveitis (C). Log LDA >2 for all taxa. * Passed FDR correction threshold. ** Passed FDR and Bonferroni correction thresholds.
Host genes determine the intestinal microbial community composition
To investigate whether other host metadata associated with the intestinal microbiota composition, we performed correlative analyses between taxonomic communities at all hierarchical levels, patient metadata, and HLA alleles. No significant differences were observed in alpha or beta diversity when subjects were grouped by gender (Supplementary Figure 5A) or BMI (Supplementary Figure 5B). Although there were statistically significant differences when subjects were grouped by race (Supplementary Figure 5C), further exploration revealed that this distinction was largely determined by HLA status.
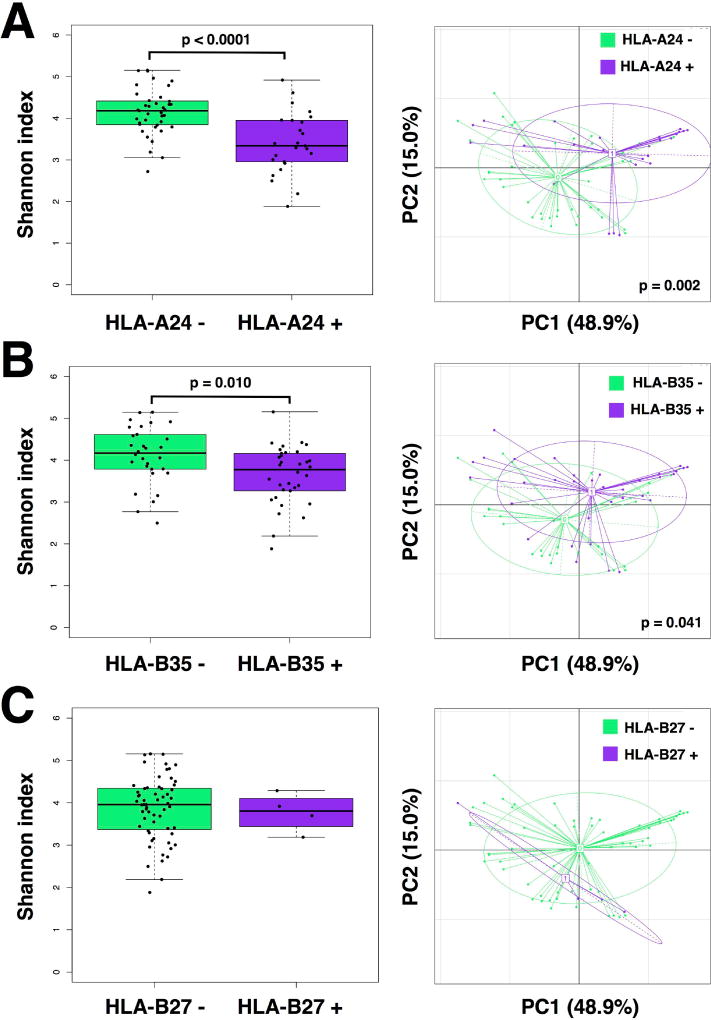
Alpha diversity was significantly lower in those carrying the HLA-A24 allele (p<0.0001; Figure 3A), as well as those positive for HLA-B35 (p=0.010; Figure 3B), irrespective of race and disease status. However, the presence of HLA-A2 (Supplementary Figure 6A) and HLA-C7 (Supplementary Figure 6B) did not correlate with differences in gut microbiota diversity. Correspondingly, differences in beta diversity were also seen in cases of HLA-A24 (p=0.002; Figure 3A) and HLA-B35 (p=0.041; Figure 3B), an effect not correlated with prior antibiotic use. Given the low prevalence of HLA-B27 and HLA-B15 positive individuals, there was not enough power to detect statistical differences in diversity for HLA-B27 (Figure 3C) or HLA-B15 (Supplementary Figure 6C). To exclude the possibility that these changes were biased by the low number of Caucasians or by antecedent GU infections, we repeated the analysis excluding Caucasians and including only GI infections, noting similar results in both cases (Supplementary Table 1).
Figure 3.
Alpha and beta diversity for subjects grouped by HLA alleles. Alpha diversity was significantly lower in subjects carrying the HLA-A24 allele (p<0.0001) (A) and the HLA-B35 allele (p=0.010) (B). Correspondingly, differences in beta diversity were also seen in cases of HLA-A24 (p=0.002) (A) and HLA-B35 (p=0.041) (B). There were too few HLA-B27 positive subjects to calculate a statistical difference (C). Alpha diversity was determined by the Shannon diversity index. Beta diversity was determined by weighted UniFrac distances. Statistical significance was calculated using the Mann-Whitney U test for alpha diversity and the Adonis (Permanova) test for beta diversity.
Interrelations of gut microbiota and metadata in ReA
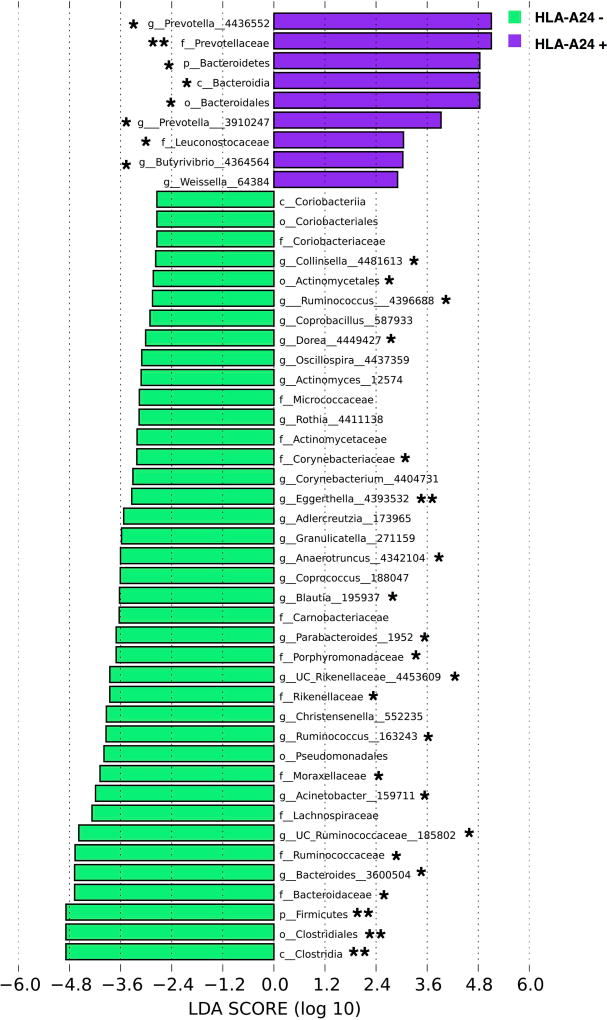
To gain a better insight into specific taxa driving these host-microbiome interactions, we examined the relationship between the gut community composition and HLA-A24 allele status. LEfSe analysis demonstrated a number of taxa that were distinct between the HLA-A24 positive and negative groups (Figure 4). Of genera that passed FDR correction threshold, the relative abundance of Prevotella was significantly higher in subjects who carried an HLA-A24 allele (Supplementary Figure 7A). At the family level, Prevotellaceae was more abundant in the HLA-A24 positive group (Supplementary Figure 7B), whereas Rikenellaceae and Ruminococcaceae were more abundant in the HLA-A24 negative group (Supplementary Figure 7C–D).
Figure 4.
LEfSe analysis of HLA-A24 positive and negative subjects. LDA demonstrates that HLA-A24 negative subjects had overall more differentiating taxa compared to HLA-A24 positive subjects. A number of genera passed FDR correction threshold. Most notably, Prevotella was enriched in subjects who had the HLA-A24 allele. Log LDA >2 for all taxa.
* Passed FDR correction threshold. ** Passed FDR and Bonferroni correction thresholds.
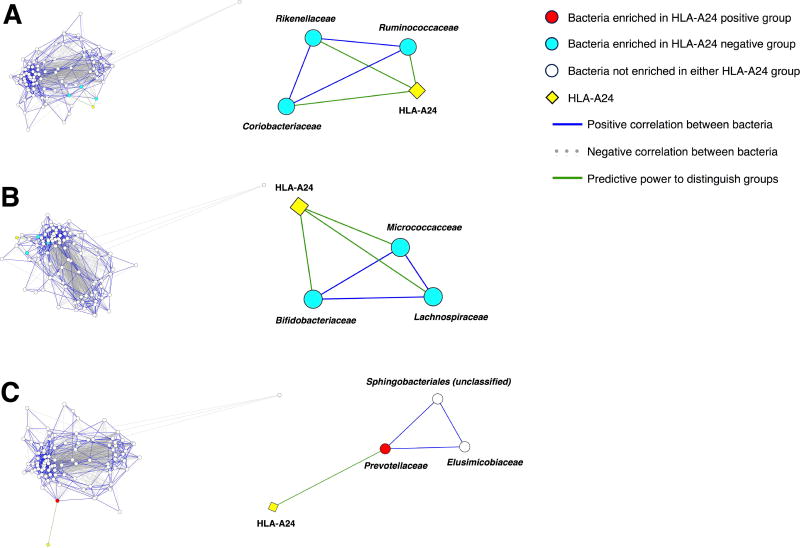
To identify groups of bacteria that might be synergistically correlated with HLA-A24, we performed network analysis to determine bacteria that co-occurred with taxa identified by LEfSe and that were also predictive of HLA-A24 status. Representative groups (maximal cliques; see Materials and Methods section) of bacteria at the family level are shown in Figure 5. Taxa in cliques comprising Ruminococcaceae-Rikenellaceae-Coriobacteriaceae (Figure 5A) and Lachnospiraceae-Micrococcaceae-Bifidobacteriaceae (Figure 5B) were all enriched in the HLA-A24 negative cohort, and each bacterium had predictive power for HLA-A24 status. In the clique comprising Prevotellaceae-Sphingobacteriales (unclassified)-Elusimicrobiaceae (Figure 5C), only Prevotellaceae was enriched in the HLA-A24 positive cohort and had predictive power for HLA-A24 status.
Figure 5.
Co-occurrence network analysis at the family level. Left side of the figure demonstrates the entire network of relationships among taxa. Right side of the figure demonstrates specific cliques of taxa, which co-occur with one another and with at least one taxon in each clique having predictive power for HLA-A24 allele status. In the overall network and cliques, edge lengths are inversely proportional to the strength of the correlation or the strength of predictive power that distinguishes between HLA-A24 positive and negative groups. Taxa that were enriched in the HLA-A24 negative cohort; each individual taxon also had predictive power for HLA-A24 status (A and B). Only Prevotellaceae was enriched in the HLA-A24 positive cohort and had predictive power for HLA-A24 status (C).
DISCUSSION
In this study, we report that the prevalence of enteropathogens in subjects with ReA and post-infectious peripheral SpA was significantly increased, while several gut commensals were decreased. Intriguingly, host genetics was a major driver for microbiota richness and diversity, irrespective of disease status.
Along with IBD-related arthritis and Whipple’s disease, ReA represents one of the clearest examples of the “gut-synovial” axis spectrum disorders. The pathogenesis of ReA, however, has been studied only in the context of culture-dependent microbiology methods. The overall consensus in the field is that intestinal pathogens, most notably Yersinia, Salmonella, Shigella, and Campylobacter, are triggering events for systemic articular inflammation (1).
Multiple groups have linked the intestinal microbiota of animals and humans to host genetics, gut inflammatory responses, and the different clinical phenotypes of SpA (12). Two decades ago Taurog and colleagues determined that luminal bacteria mediated the development of arthritis and colitis in HLA-B27 transgenic rats (30, 31). Since then, many studies have contributed findings corroborating this observation. Work investigating genetically susceptible animals have shown that gut dysbiosis activates a Th17-mediated immune response in the host’s intestinal lamina propria, thereby promoting local and systemic inflammation in the form of joint disease (32, 33). Clinically overt and subclinical histologic and molecular markers of gut inflammation have also been found in patients with ankylosing spondylitis (AS) (34) and in those with PsA (35). Furthermore, a recent study demonstrated that an outer membrane protein of Salmonella typhimurium can induce synovial fluid mononuclear cells in patients with ReA to upregulate cytokines associated with the IL-23/IL-17 axis (36).
This was followed by several communications describing a dysbiotic process in patients with new-onset RA (37), animal models of SpA (38, 39), and patient cohorts with SpA, including AS (40, 41) and PsA (29). A summary of these findings in humans and animal models can be found in a recent review by our group (42). In line with these data, our research demonstrates that subjects with ReA were characterized as having a decreased abundance of commensals in their gut microbiota compared to controls, which were enriched in the family Lachnospiraceae with associated genera—Blautia, Coprococcus, Roseburia, as well as the genus Collinsella. Interestingly, there was a concomitant increase in the prevalence of enteropathogens in the ReA group, particularly Pseudomonas and Erwinia, a taxon that has ≥97% identity with the usual ReA-associated gut bacteria.
To our knowledge, this is the first culture-independent, high-throughput, DNA sequencing study characterizing the overall gut community composition in post-infectious peripheral SpA. Prior efforts were directed towards only a handful of culturable organisms (1), and did not report on the presence or abundance of intestinal commensals. Our findings suggest that even when the overall bacterial composition is not significantly different from controls, a decrease in several potentially protective bacteria could prime genetically susceptible individuals with GI or GU infections to develop ReA. This contrasts with subjects who eventually contain the immune response to the lamina propria due to the prevalence of commensals. Intriguingly, two of the underrepresented commensal taxa in ReA—Lachnospiraceae and Coprococcus— were also decreased in patients with new-onset PsA (29). Similarly, we found a positive correlation between the presence of the genus Dialister and the development of both uveitis and sacroiliitis. This same genus has recently been reported as a marker of disease activity in AS (41). Since ReA, AS, and PsA all belong to the SpA spectrum of diseases, it is plausible that a perturbation in the intestinal microbiota constitutes yet another common environmental risk factor among patients with these disorders. Whether the perturbations are causative or a consequence of SpA-related inflammation remains to be elucidated.
Although the relative abundance of the usual enteropathogens associated with post-infectious peripheral SpA was expectedly low (less than 1% abundance for each microorganism), there was a significantly higher prevalence of such taxa in ReA subjects compared to controls, as seen in other autoimmune conditions such as IBD (43) and further validating prior culture-dependent observations. Interestingly, the majority of ReA cases were reported in the months of April–July (28). This seasonal effect could be a determinant of disease development as it may correlate with differences in the types of pathogens that are present.
Perhaps even more remarkable is the observation that certain HLA alleles seemed to determine microbiota composition in both health and disease. These effects were independent of either disease status or prior antibiotic use. A similar finding was reported in an RA-like murine model, where HLA transgenic animals carrying the DRB1*0401 gene were more susceptible to collagen-induced arthritis. These mice showed gut microbiome dysbiosis and enrichment with Clostridium-like bacteria (44). In our study, there were significant differences in intestinal community diversity and species composition associated with HLA-A24 and, to a lesser degree, with HLA-B35. Such differences, however, were not seen with the HLA-B27 allele, which was present in only a minority of subjects. This was not surprising since prior work indicates that only 4% of Guatemalan patients with SpA are HLA-B27 positive (10). On the other hand, the frequency of the HLA-A24 allele in Amerindian populations is quite elevated and estimated to be >20% (45). The presence of this allele has also been associated with a number of autoimmune diseases in non-Caucasians, including type 1 diabetes in Japan (46) and RA in a Moroccan cohort (47). Most relevant to this study, HLA-A*2402 also correlated with AS in a Basque cohort, independent of HLA-B27 status (48). Multiple lines of evidence therefore suggest that microbial-host gene interactions conferring susceptibility for autoimmune disease can significantly vary among ethnic populations, particularly with respect to HLA alleles.
Surveying the microbiota composition, Prevotella (genus) and Prevotellaceae (family) were more abundant in subjects who were HLA-A24 positive. Prevotella has been implicated in the pathogenesis of RA. In particular, Prevotella copri correlated with disease in new-onset rheumatoid arthritis (NORA) patients, while the relative abundance of P. copri was lower in subjects who had the DRB1 shared-epitope RA risk alleles (37). This suggests that a certain threshold for P. copri abundance may be necessary to overcome the lack of genetic predisposition in the case of RA subjects. In the case of ReA, it is possible that Prevotella-HLA-24 co-occurrence is necessary but insufficient as a triggering factor for post-infectious arthritis unless there is a concomitant reduction in beneficial, gut-protective commensals.
Interestingly, Ruminococcaceae and Rikenellaceae were more abundant in HLA-A24 negative subjects. Furthermore, Ruminococcaceae and Rikenellaceae co-occurred with one another, and each had predictive power for HLA-A24 status. Studies of HLA-B27 transgenic rats that exhibit many features of SpA also revealed a higher abundance of Prevotella and a lower abundance of Rikenellaceae (38). Moreover, terminal ileum biopsies from AS subjects had a higher abundance of Ruminococcaceae and Rikenellaceae, and a lower abundance of Prevotellaceae (40). Although this data could represent a true correlation with disease status, co-occurrence with host genetics was not performed since all AS participants were HLA-B27 positive.
Taken together, our findings further support the possibility that host genetics can prime gut microbiome features to trigger autoimmunity in susceptible individuals. We propose that moving forward, the characterization of these gene-microbiome interactions should be an integral part of any correlative human studies to avoid potential confounders that could implicate taxa to disease processes.
We acknowledge several limitations of our study. First, there was a relatively small number of participants and geographic/ethnic homogeneity, rendering the results less generalizable to other populations. However, this could also represent a strength since this is a cohort that has not been previously evaluated, offering a new outlook on ReA pathogenesis, especially with respect to host genetics. Second, our study did not incorporate microbiota of controls without preceding GI or GU infections. Although we are not aware of any studies that have characterized the bacterial communities of the healthy Native and Mixed populations in Guatemala, a recent study of uncontacted Amerindians in Venezuela showed high abundance of Prevotella and low abundance of Bacteroides (49).
Another limitation relates to the lack of timely and culture-proven diagnosis of preceding infections, which is a widely acknowledged challenge in research ReA studies and in most cases of ReA diagnosed in the clinical setting (50). In fact, stool cultures can occasionally confirm a preceding or concomitant infection with a classic ReA pathogen, but by the time subjects develop arthritis, their symptoms have usually resolved and the pathogens are no longer retrievable. Furthermore, serological tests are non-specific and are only able to demonstrate prior immunity rather than causality. For these reasons, we relied on subject-reported history of antecedent infection, but acknowledge that some of the infections could have been viral or secondary to other etiologies (i.e., post-infectious peripheral SpA being a more precise definition of our patient population). We also detected enteropathogens in the ReA cohort of our study, only implying persistence of an organism’s DNA, as 16S rDNA sequencing studies do not allow for viability determination. However, this may be of relevance since the pathogenesis of ReA is yet unknown, and some hypotheses suggest that microbial components may drive the downstream inflammatory response (12).
Uveitis was another self-reported variable and it is possible that some of the cases were actually conjunctivitis or other related conditions. Nevertheless, 75% of the cases were seen by an ophthalmologist, deemed to have uveitis, and subsequently referred to a rheumatology clinic so misdiagnosis is less likely. We also note that incidence of sacroiliitis was unexpectedly high in both the arthritis and control groups. Because most of them of were adjudicated as grade I sacroiliitis, it is possible that this could represent over-reporting, which is also quite common in clinical practice. Unfortunately, we could not pursue further blinding or centralized readings for confirmatory purposes and therefore these findings should be interpreted with caution.
The use of antibiotics for the treatment of infections prior to sample collection may have also had a dysbiotic effect on the gut microbiome. However, our results were independent of antibiotic use. In the future, baseline and post-arthritis characterization of microbial communities in at-risk populations may help discern these perturbations in a prospective fashion. Complementary studies utilizing metagenomics and metabolomics platforms will also be required to better understand the role of gut bacterial enzymes and their byproducts in ReA pathogenesis.
In summary, this study further enhances our understanding of ReA and post-infectious peripheral SpA in a non-HLA-B27 dominated population. Utilizing a high-throughput DNA sequencing approach, we identified several organisms that are differentially present in samples from ReA subjects compared to controls. These taxa and the lack of specific commensals, as well as the associated changes in immune response, will require further research and validation in other cohorts. The effect of host genetics on shaping the human microbiome in health and disease is also intriguing, and could shed light on our understanding of the symbiotic relationship between host and community diversity and composition. Furthermore, it is plausible that the combination of exposure to known pathogens, host genetics, and the makeup of commensal microorganisms could confer susceptibility for the development of ReA, post-infectious peripheral SpA and other variants of the SpA-spectrum disorders. Future mechanistic research coupled with well-designed prospective human cohorts are needed to close these knowledge gaps in the field.
Supplementary Material
Supplementary Figure 1: Microbiota alpha and beta diversity in reactive arthritis (ReA) and control (Ctrl) subjects. Alpha diversity was determined by the number of unique OTUs (A), the Shannon diversity index (B), and the Faith’s phylodiversity index (C). None were statistically significant, but there was an overall trend toward decreased diversity in ReA subjects with the Shannon diversity index. Beta diversity, as determined by weighted UniFrac distances, showed no distinct clustering patterns (D). Statistical significance was calculated using the Mann-Whitney U test for alpha diversity and the Adonis (Permanova) test for beta diversity.
Supplementary Figure 2: Alpha and beta diversity in subjects grouped by antibiotic (Abx) and sulfasalazine (SSA) exposure. No statistically significant differences in alpha or beta diversity were seen when subjects were grouped by whether they received antibiotics (A). Too few subjects were exposed to SSA to detect a statistical difference (B). Alpha diversity was determined by the Shannon diversity index. Beta diversity was determined by weighted UniFrac distances. Statistical significance was calculated using the Mann-Whitney U test for alpha diversity and the Adonis (Permanova) test for beta diversity.
Supplementary Figure 3: Prevalence of usual intestinal enteropathogens in reactive arthritis (ReA) and control (Ctrl) subjects. Bacteria include Salmonella, Shigella, Campylobacter, and Erwinia (which has ≥97% identity to various species of Salmonella, Shigella, and Yersinia using a nucleotide BLAST search). Each box represents a single individual. The darker the intensity of the box, the greater the number of usual enteropathogens present. At least one enteropathogen was present in 71.9% of ReA subjects vs 46.8% of control subjects (p=0.042). Each enteropathogen constituted less than 1% of bacterial relative abundance (data not shown). Statistical significance was calculated using the chi-square test.
Supplementary Figure 4: Alpha and beta diversity for clinical and radiographic metadata specific to reactive arthritis. Alpha diversity was significantly lower in subjects with ultrasound evidence of enthesitis (A). Beta diversity was significantly different in subjects with radiographic evidence of sacroiliitis (B). No differences in alpha or beta diversity were seen in subjects with uveitis (C). Alpha diversity was determined by the Shannon diversity index. Beta diversity was determined by weighted UniFrac distances. Statistical significance was calculated using the Mann-Whitney U test for alpha diversity and the Adonis (Permanova) test for beta diversity.
Supplementary Figure 5: Alpha and beta diversity for sex, BMI, and race. There were no significant differences in alpha or beta diversity when subjects were grouped by sex (A) or BMI (B). BMI was categorized by normal: 18.5–25.0 kg/m2, overweight: 25.1–30.0 kg/m2, and obese: >30 kg/m2. One subject <18.5 kg/m2 was included in the normal group. There were statistically significant differences in alpha and beta diversity when subjects were grouped by race, particularly with regard to Caucasian (Blanca) compared to the Native (Indigena) and Mixed (Mestiza) populations (C). However, this distinction was largely attributable to HLA status. Alpha diversity was determined by the Shannon diversity index. Beta diversity was determined by weighted UniFrac distances. Statistical significance was calculated using the Mann-Whitney U and Kruskal-Wallis tests for alpha diversity and the Adonis (Permanova) test for beta diversity.
Supplementary Figure 6: Alpha and beta diversity for subjects grouped by HLA alleles. There were no statistically significant differences in alpha or beta diversity when subjects were grouped by HLA-A2 (A) or HLA-C7 (B) status. There were too few HLA-B15 positive subjects to calculate a statistical difference (C). Alpha diversity was determined by the Shannon diversity index. Beta diversity was determined by weighted UniFrac distances. Statistical significance was calculated using the Mann-Whitney U test for alpha diversity and the Adonis (Permanova) test for beta diversity.
Supplementary Figure 7: Relative abundances of certain taxa that passed FDR correction threshold in LEfSe analysis of HLA-A24 positive and negative subjects. On the genus level, Prevotella relative abundance was higher in HLA-A24 positive subjects (A). On the family level, Prevotellaceae was more abundant in the HLA-A24 positive group (B), whereas Rikenellaceae and Ruminococcaceae were more abundant in the HLA-A24 negative group (C–D). Statistical significance was calculated using the Mann-Whitney U test.
Supplementary Table 1: Comparison of p-values for alpha and beta diversity across all study parameters among three groups: Group 1—all subjects (left column), Group 2—Caucasian subjects excluded from analysis (middle column), Group 3—only subjects with gastrointestinal (GI) infection included in analysis (right column). There were very few differences among the three groups with the exception of race, which showed statistically significant differences in alpha and beta diversity for Groups 1 and 3 due to the presence of Caucasian subjects who were absent in Group 2. Alpha diversity was determined by the Shannon diversity index. Beta diversity was determined by weighted UniFrac distances. Statistical significance was calculated using the Mann-Whitney U test or Kruskal-Wallis test for alpha diversity and the Adonis (Permanova) test for beta diversity. * Not enough power to detect a statistical difference.
Table 2.
Spondyloarthropathy Characteristics
| Variables | Patients with ReA (n=32) |
|---|---|
| History of Peripheral Arthritis n (%) | 32 (100 %) |
| History of Sero-Negative SpAs n (%) | 3 (9 %) |
| First-Degree Relatives with SpAs n (%) | 2 (6 %) |
| Symptoms n (%) | |
| Back pain | 29 (91 %) |
| Uveitis | 20 (63 %) |
| SpA Medications n (%) | |
| NSAIDs | 31 (97 %) |
| Glucocorticoids | 2 (6 %) |
| Medicinal Herbs | 10 (31 %) |
| Abnormal Imaging (?*) | |
| Ultrasound Achilles Enthesopathy | 14 (44 %) |
| Tender Joints n (%) | |
| Right Sacroiliac | 27 (82 %) |
| Left Sacroiliac | 25 (76 %) |
| Right Ankle | 16 (49 %) |
| Left Ankle | 18 (55 %) |
| Right Elbow | 18 (55 %) |
| Left Elbow | 10 (30%) |
| Right Knee | 7 (21%) |
| Left Knee | 9 (27%) |
| Right Shoulder | 16 (50%) |
| Left Shoulder | 6 (18%) |
| Right Wrist | 4 (12%) |
| Left Wrist | 3 (9%) |
| Right Distal Interphalangeal | 0 (0%) |
| Left Distal Interphalangeal | 0 (0%) |
| Right Proximal Interphalangeal | 2 (6%) |
| Left Proximal Interphalangeal | 1 (3%) |
| Right Metacarpal | 5 (15%) |
| Left Metacarpal | 2 (6%) |
| Right Metatarsal | 1 (3%) |
| Swollen Joints n (%) | |
| Right Ankle | 3 (9 %) |
| Left Ankle | 4 (13 %) |
| Right Elbow | 2 (6 %) |
| Left Elbow | 3 (9%) |
| Right Knee | 1 (3%) |
| Left Knee | 1 (3%) |
| Right Wrist | 2 (6%) |
| Left Wrist | 2 (6%) |
| Right Proximal Interphalangeal | 1 (3%) |
| Left Metacarpal | 1 (3%) |
| Tender Entheses | |
| Right Achilles Tendon | 22 (69 %) |
| Left Achilles Tendon | 22 (69 %) |
| Right Medial Femoral Condyle | 17 (53 %) |
| Left Medial Femoral Condyle | 16 (50%) |
| Right Lateral Femoral Condyle | 7 (22%) |
| Left Lateral Femoral Condyle | 7 (22%) |
| Right Plantar Fascia | 16 (50%) |
| Left Plantar Fascia | 15 (47%) |
Most controls did not report any SpA related symptoms, medication use, joint tenderness, or enthesopathy with the exception of 6% reporting uveitis, 16% reporting herbal medication use, and 6% found to have abnormal ultrasound Achilles enthesopathy imaging.
Acknowledgments
We would like to thank Parvathy Girija for extracting DNA from fecal samples and preparing them for sequencing analysis. We would also like to thank Benjamin Wu for his advice and assistance during the data analysis phase of this project.
Supported by:
Guatemala-Penn Partners Program and NIH Fogarty International Center grant D43TW008317; Grant No. K23 AR063764 from NIAMS and the Rheumatology Research Foundation to Dr. Ogdie; Grants No. K23 AR064318 and R03 AR072182 from NIAMS to Dr. Scher; The Colton Center for Autoimmunity; The Riley Family Foundation; The Snyder Family Foundation
References
- 1.Hannu T. Reactive arthritis. Best Pract Res Clin Rheumatol. 2011;25(3):347–57. doi: 10.1016/j.berh.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 2.Schiellerup P, Krogfelt KA, Locht H. A comparison of self-reported joint symptoms following infection with different enteric pathogens: effect of HLA-B27. J Rheumatol. 2008;35(3):480–7. [PubMed] [Google Scholar]
- 3.Putterman C, Rubinow A. Reactive arthritis associated with Clostridium difficile pseudomembranous colitis. Semin Arthritis Rheum. 1993;22(6):420–6. doi: 10.1016/s0049-0172(05)80033-2. [DOI] [PubMed] [Google Scholar]
- 4.Brewerton DA, Caffrey M, Nicholls A, Walters D, Oates JK, James DC. Reiter's disease and HL-A 27. Lancet. 1973;302(7836):996–8. doi: 10.1016/s0140-6736(73)91091-x. [DOI] [PubMed] [Google Scholar]
- 5.Khan MA. HLA-B27 and its subtypes in world populations. Curr Opin Rheumatol. 1995;7(4):263–9. doi: 10.1097/00002281-199507000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Sieper J, Rudwaleit M, Braun J, van der Heijde D. Diagnosing reactive arthritis: role of clinical setting in the value of serologic and microbiologic assays. Arthritis Rheum. 2002;46(2):319–27. doi: 10.1002/art.504. [DOI] [PubMed] [Google Scholar]
- 7.Carter JD, Hudson AP. Reactive arthritis: clinical aspects and medical management. Rheum Dis Clin North Am. 2009;35(1):21–44. doi: 10.1016/j.rdc.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Townes JM, Deodhar AA, Laine ES, Smith K, Krug HE, Barkhuizen A, et al. Reactive arthritis following culture-confirmed infections with bacterial enteric pathogens in Minnesota and Oregon: a population-based study. Ann Rheum Dis. 2008;67(12):1689–96. doi: 10.1136/ard.2007.083451. [DOI] [PubMed] [Google Scholar]
- 9.Solitar BM, Lozada CJ, Tseng CE, Lowe AM, Krajewski WM, Blanchard K, et al. Reiter's syndrome among Asian shipboard immigrants: the case of The Golden Venture. Semin Arthritis Rheum. 1998;27(5):293–300. doi: 10.1016/s0049-0172(98)80050-4. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Kutzbach A, Montenegro A, Iraheta I, Bara C, Saenz R. Epidemiology of spondyloarthropathies in Central America. Am J Med Sci. 2011;341(4):295–7. doi: 10.1097/MAJ.0b013e31820f8cf1. [DOI] [PubMed] [Google Scholar]
- 11.Tsui FW, Xi N, Rohekar S, Riarh R, Bilotta R, Tsui HW, et al. Toll-like receptor 2 variants are associated with acute reactive arthritis. Arthritis Rheum. 2008;58(11):3436–8. doi: 10.1002/art.23967. [DOI] [PubMed] [Google Scholar]
- 12.Manasson J, Scher JU. Spondyloarthritis and the microbiome: new insights from an ancient hypothesis. Curr Rheumatol Rep. 2015;17(2):10. doi: 10.1007/s11926-014-0487-7. [DOI] [PubMed] [Google Scholar]
- 13.Siala M, Gdoura R, Fourati H, Rihl M, Jaulhac B, Younes M, et al. Broad-range PCR, cloning and sequencing of the full 16S rRNA gene for detection of bacterial DNA in synovial fluid samples of Tunisian patients with reactive and undifferentiated arthritis. Arthritis Res Ther. 2009;11(4):R102. doi: 10.1186/ar2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granfors K, Jalkanen S, Lindberg AA, Maki-Ikola O, von Essen R, Lahesmaa-Rantala R, et al. Salmonella lipopolysaccharide in synovial cells from patients with reactive arthritis. Lancet. 1990;335(8691):685–8. doi: 10.1016/0140-6736(90)90804-e. [DOI] [PubMed] [Google Scholar]
- 15.Paniagua-Avila MA, Messenger E, Nelson CA, Calgua E, Barg FK, Bream KW, et al. The Guatemala-Penn Partners: An Innovative Inter-Institutional Model for Scientific Capacity-Building, Healthcare Education, and Public Health. Front Public Health. 2017;5:70. doi: 10.3389/fpubh.2017.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudwaleit M, van der Heijde D, Landewe R, Akkoc N, Brandt J, Chou CT, et al. The Assessment of SpondyloArthritis International Society classification criteria for peripheral spondyloarthritis and for spondyloarthritis in general. Ann Rheum Dis. 2011;70(1):25–31. doi: 10.1136/ard.2010.133645. [DOI] [PubMed] [Google Scholar]
- 17.Scher JU, Joshua V, Artacho A, Abdollahi-Roodsaz S, Ockinger J, Kullberg S, et al. The lung microbiota in early rheumatoid arthritis and autoimmunity. Microbiome. 2016;4(1):60. doi: 10.1186/s40168-016-0206-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 20.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8(9):e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bron C, Kerbosch J. Finding All Cliques of an Undirected Graph [H] Commun Acm. 1973;16(9):575–7. [Google Scholar]
- 26.Hagberg A, Schult D, Swart P. Exploring Network Structure, Dynamics, and Function using NetworkX. In: Varoquaux G, Vaught T, Millman J, editors. 7th Python in Science conference (SciPy 2008); Pasadena, CA, USA. 2008. pp. 11–5. [Google Scholar]
- 27.GraphPad Prism. 7.0a for Mac ed. San Diego, CA: GraphPad Software; [Google Scholar]
- 28.Garcia Ferrer HR, Garcia Kutzbach A, Iraheta I, Scher JU, Von Feldt J, Ogdie-Beatty A. Clinical and Imaging Features of Reactive Arthritis in Guatemala City [abstract] Arthritis Rheumatol. 2015;67(suppl 10) http://acrabstracts.org/abstract/clinical-and-imaging-features-of-reactive-arthritis-in-guatemala-city/ [Google Scholar]
- 29.Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67(1):128–39. doi: 10.1002/art.38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rath HC, Herfarth HH, Ikeda JS, Grenther WB, Hamm TE, Jr, Balish E, et al. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Invest. 1996;98(4):945–53. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernandez-Sueiro JL, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180(6):2359–64. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–27. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdollahi-Roodsaz S, Joosten LA, Koenders MI, Devesa I, Roelofs MF, Radstake TR, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest. 2008;118(1):205–16. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mielants H, Veys EM, Cuvelier C, De Vos M, Goemaere S, De Clercq L, et al. The evolution of spondyloarthropathies in relation to gut histology. II. Histological aspects. J Rheumatol. 1995;22(12):2273–8. [PubMed] [Google Scholar]
- 35.Ciccia F, Guggino G, Ferrante A, Raimondo S, Bignone R, Rodolico V, et al. Interleukin-9 Overexpression and Th9 Polarization Characterize the Inflamed Gut, the Synovial Tissue, and the Peripheral Blood of Patients With Psoriatic Arthritis. Arthritis Rheumatol. 2016;68(8):1922–31. doi: 10.1002/art.39649. [DOI] [PubMed] [Google Scholar]
- 36.Chaurasia S, Shasany AK, Aggarwal A, Misra R. Recombinant Salmonella typhimurium outer membrane protein A is recognized by synovial fluid CD8 cells and stimulates synovial fluid mononuclear cells to produce interleukin (IL)-17/IL-23 in patients with reactive arthritis and undifferentiated spondyloarthropathy. Clin Exp Immunol. 2016;185(2):210–8. doi: 10.1111/cei.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin P, Bach M, Asquith M, Lee AY, Akileswaran L, Stauffer P, et al. HLA-B27 and human beta2-microglobulin affect the gut microbiota of transgenic rats. PLoS One. 2014;9(8):e105684. doi: 10.1371/journal.pone.0105684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asquith MJ, Stauffer P, Davin S, Mitchell C, Lin P, Rosenbaum JT. Perturbed Mucosal Immunity and Dysbiosis Accompany Clinical Disease in a Rat Model of Spondyloarthritis. Arthritis Rheumatol. 2016;68(9):2151–62. doi: 10.1002/art.39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costello ME, Ciccia F, Willner D, Warrington N, Robinson PC, Gardiner B, et al. Brief Report: Intestinal Dysbiosis in Ankylosing Spondylitis. Arthritis Rheumatol. 2015;67(3):686–91. doi: 10.1002/art.38967. [DOI] [PubMed] [Google Scholar]
- 41.Tito RY, Cypers H, Joossens M, Varkas G, Van Praet L, Glorieus E, et al. Brief Report: Dialister as a Microbial Marker of Disease Activity in Spondyloarthritis. Arthritis Rheumatol. 2017;69(1):114–21. doi: 10.1002/art.39802. [DOI] [PubMed] [Google Scholar]
- 42.Scher JU, Littman DR, Abramson SB. Microbiome in Inflammatory Arthritis and Human Rheumatic Diseases. Arthritis Rheumatol. 2016;68(1):35–45. doi: 10.1002/art.39259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Medina M, Garcia-Gil LJ. Escherichia coli in chronic inflammatory bowel diseases: An update on adherent invasive Escherichia coli pathogenicity. World J Gastrointest Pathophysiol. 2014;5(3):213–27. doi: 10.4291/wjgp.v5.i3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez A, Luckey D, Yeoman CJ, Marietta EV, Berg Miller ME, Murray JA, et al. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS One. 2012;7(4):e36095. doi: 10.1371/journal.pone.0036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43(Database issue):D784–8. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakanishi K, Inoko H. Combination of HLA-A24, -DQA1*03, and -DR9 contributes to acute-onset and early complete beta-cell destruction in type 1 diabetes: longitudinal study of residual beta-cell function. Diabetes. 2006;55(6):1862–8. doi: 10.2337/db05-1049. [DOI] [PubMed] [Google Scholar]
- 47.Atouf O, Benbouazza K, Brick C, Bzami F, Bennani N, Amine B, et al. HLA polymorphism and early rheumatoid arthritis in the Moroccan population. Joint Bone Spine. 2008;75(5):554–8. doi: 10.1016/j.jbspin.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 48.de Juan MD, Reta A, Belzunegui J, Figueroa M, Maruri N, Cuadrado E. HLA-A*2402 and a microsatellite (D6S248) are secondary independent susceptibility markers to ankylosing spondylitis in Basque patients. Hum Immunol. 2004;65(2):175–80. doi: 10.1016/j.humimm.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1(3) doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hannu T, Inman R, Granfors K, Leirisalo-Repo M. Reactive arthritis or post-infectious arthritis? Best Pract Res Clin Rheumatol. 2006;20(3):419–33. doi: 10.1016/j.berh.2006.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Microbiota alpha and beta diversity in reactive arthritis (ReA) and control (Ctrl) subjects. Alpha diversity was determined by the number of unique OTUs (A), the Shannon diversity index (B), and the Faith’s phylodiversity index (C). None were statistically significant, but there was an overall trend toward decreased diversity in ReA subjects with the Shannon diversity index. Beta diversity, as determined by weighted UniFrac distances, showed no distinct clustering patterns (D). Statistical significance was calculated using the Mann-Whitney U test for alpha diversity and the Adonis (Permanova) test for beta diversity.
Supplementary Figure 2: Alpha and beta diversity in subjects grouped by antibiotic (Abx) and sulfasalazine (SSA) exposure. No statistically significant differences in alpha or beta diversity were seen when subjects were grouped by whether they received antibiotics (A). Too few subjects were exposed to SSA to detect a statistical difference (B). Alpha diversity was determined by the Shannon diversity index. Beta diversity was determined by weighted UniFrac distances. Statistical significance was calculated using the Mann-Whitney U test for alpha diversity and the Adonis (Permanova) test for beta diversity.
Supplementary Figure 3: Prevalence of usual intestinal enteropathogens in reactive arthritis (ReA) and control (Ctrl) subjects. Bacteria include Salmonella, Shigella, Campylobacter, and Erwinia (which has ≥97% identity to various species of Salmonella, Shigella, and Yersinia using a nucleotide BLAST search). Each box represents a single individual. The darker the intensity of the box, the greater the number of usual enteropathogens present. At least one enteropathogen was present in 71.9% of ReA subjects vs 46.8% of control subjects (p=0.042). Each enteropathogen constituted less than 1% of bacterial relative abundance (data not shown). Statistical significance was calculated using the chi-square test.
Supplementary Figure 4: Alpha and beta diversity for clinical and radiographic metadata specific to reactive arthritis. Alpha diversity was significantly lower in subjects with ultrasound evidence of enthesitis (A). Beta diversity was significantly different in subjects with radiographic evidence of sacroiliitis (B). No differences in alpha or beta diversity were seen in subjects with uveitis (C). Alpha diversity was determined by the Shannon diversity index. Beta diversity was determined by weighted UniFrac distances. Statistical significance was calculated using the Mann-Whitney U test for alpha diversity and the Adonis (Permanova) test for beta diversity.
Supplementary Figure 5: Alpha and beta diversity for sex, BMI, and race. There were no significant differences in alpha or beta diversity when subjects were grouped by sex (A) or BMI (B). BMI was categorized by normal: 18.5–25.0 kg/m2, overweight: 25.1–30.0 kg/m2, and obese: >30 kg/m2. One subject <18.5 kg/m2 was included in the normal group. There were statistically significant differences in alpha and beta diversity when subjects were grouped by race, particularly with regard to Caucasian (Blanca) compared to the Native (Indigena) and Mixed (Mestiza) populations (C). However, this distinction was largely attributable to HLA status. Alpha diversity was determined by the Shannon diversity index. Beta diversity was determined by weighted UniFrac distances. Statistical significance was calculated using the Mann-Whitney U and Kruskal-Wallis tests for alpha diversity and the Adonis (Permanova) test for beta diversity.
Supplementary Figure 6: Alpha and beta diversity for subjects grouped by HLA alleles. There were no statistically significant differences in alpha or beta diversity when subjects were grouped by HLA-A2 (A) or HLA-C7 (B) status. There were too few HLA-B15 positive subjects to calculate a statistical difference (C). Alpha diversity was determined by the Shannon diversity index. Beta diversity was determined by weighted UniFrac distances. Statistical significance was calculated using the Mann-Whitney U test for alpha diversity and the Adonis (Permanova) test for beta diversity.
Supplementary Figure 7: Relative abundances of certain taxa that passed FDR correction threshold in LEfSe analysis of HLA-A24 positive and negative subjects. On the genus level, Prevotella relative abundance was higher in HLA-A24 positive subjects (A). On the family level, Prevotellaceae was more abundant in the HLA-A24 positive group (B), whereas Rikenellaceae and Ruminococcaceae were more abundant in the HLA-A24 negative group (C–D). Statistical significance was calculated using the Mann-Whitney U test.
Supplementary Table 1: Comparison of p-values for alpha and beta diversity across all study parameters among three groups: Group 1—all subjects (left column), Group 2—Caucasian subjects excluded from analysis (middle column), Group 3—only subjects with gastrointestinal (GI) infection included in analysis (right column). There were very few differences among the three groups with the exception of race, which showed statistically significant differences in alpha and beta diversity for Groups 1 and 3 due to the presence of Caucasian subjects who were absent in Group 2. Alpha diversity was determined by the Shannon diversity index. Beta diversity was determined by weighted UniFrac distances. Statistical significance was calculated using the Mann-Whitney U test or Kruskal-Wallis test for alpha diversity and the Adonis (Permanova) test for beta diversity. * Not enough power to detect a statistical difference.