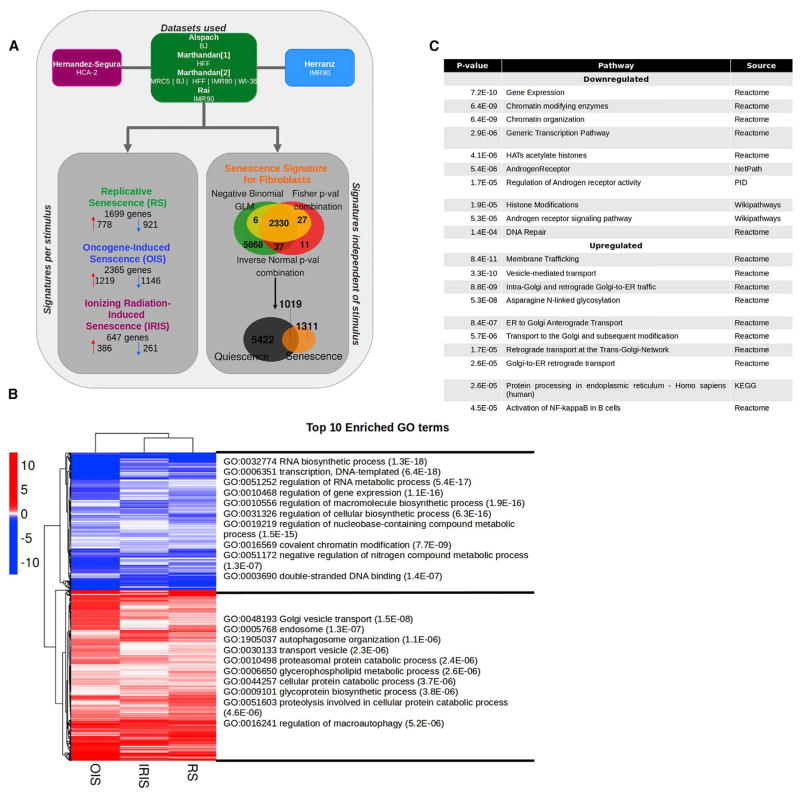
Figure 1. Meta-Analysis of Senescent Fibroblast Transcriptomics.
(A) Experimental design. Seven RNA-seq datasets obtained from the indicated studies were used to build a stimulus-specific signature and general signature of senescent fibroblasts, regardless of the stimulus. Only genes with a p ≤ 0.01, calculated by the three methods and with expression unchanged or in the opposite direction in quiescence, were included in each signature. The number of genes constituting each signature is displayed in the flower plot.
(B) Heatmap of the 1,311 genes in the senescence signature of fibroblasts and the top 10 enriched GO terms. The graph shows the logarithm base 2 of the fold change for each senescence-inducing stimulus tested with respect to proliferating cells. Blue indicates downregulated genes; red indicates upregulated genes.
(C) Top 10 enriched pathways in the senescence signature of fibroblasts. The pathways enriched in genes within the senescence signature of fibroblasts (in B) are enlisted with their corresponding p value and source. ER, endoplasmic reticulum. HAT, histone acetyltransferases; KEGG, Kyoto Encylopedia of Genes and Genomes; PID, Pathway Interaction Database.