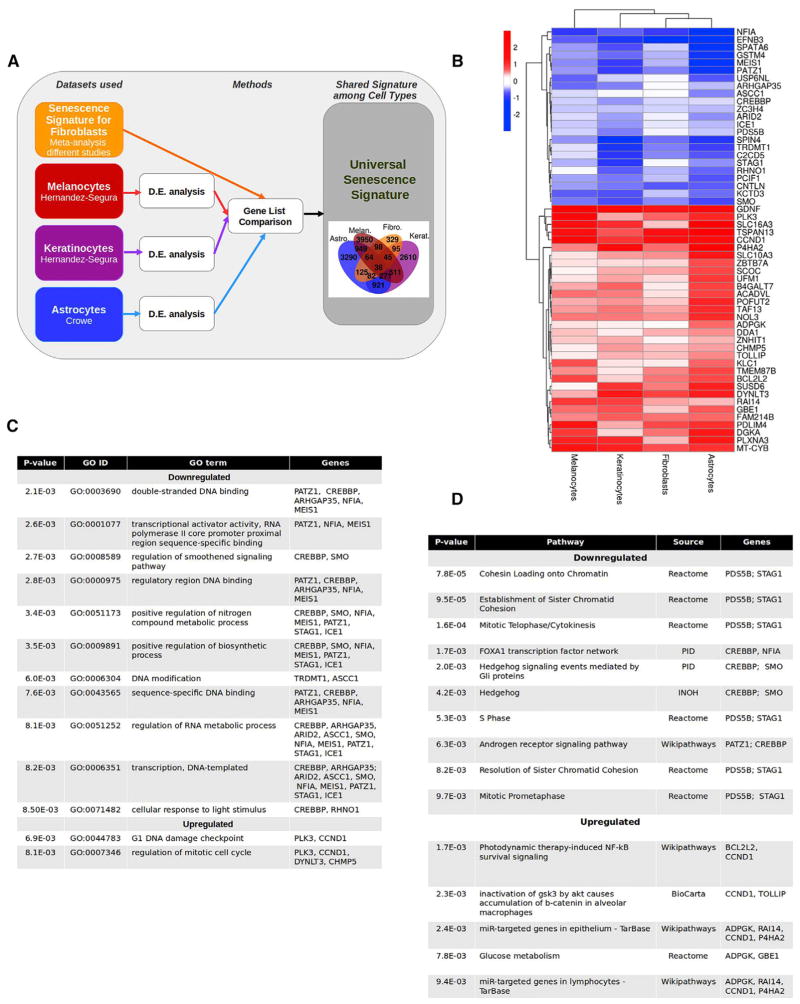
Figure 2. Characteristics of the Core Senescence-Associated Signature.
(A) Experimental design. RNA-seq datasets obtained from the indicated studies of melanocytes, keratinocytes, and astrocytes were compared to the senescence signature of fibroblasts. The intersection of genes differentially expressed (p ≤ 0.01) in all the datasets are shown in the flower plot. D.E., differential expression.
(B) Heatmap of the 55 genes of the senescence core signature. The figure shows the logarithm base 2 of the fold change for each cell type with respect to proliferating cells.
(C) GO terms enriched in the core senescence signature. The GO terms enriched in genes within the core senescence signature (in B) are listed with the corresponding p value and the associated genes.
(D) Pathways enriched in the core signature of senescence. The pathways enriched in genes within the core senescence signature (in B) are listed with their corresponding p value, source, and the associated genes. INOH, integrating network objects with hierarchies; NF-κB, nuclear factor κB; PID, Pathway Interaction Database.
See also Figures S2 and S3 and Data S2.