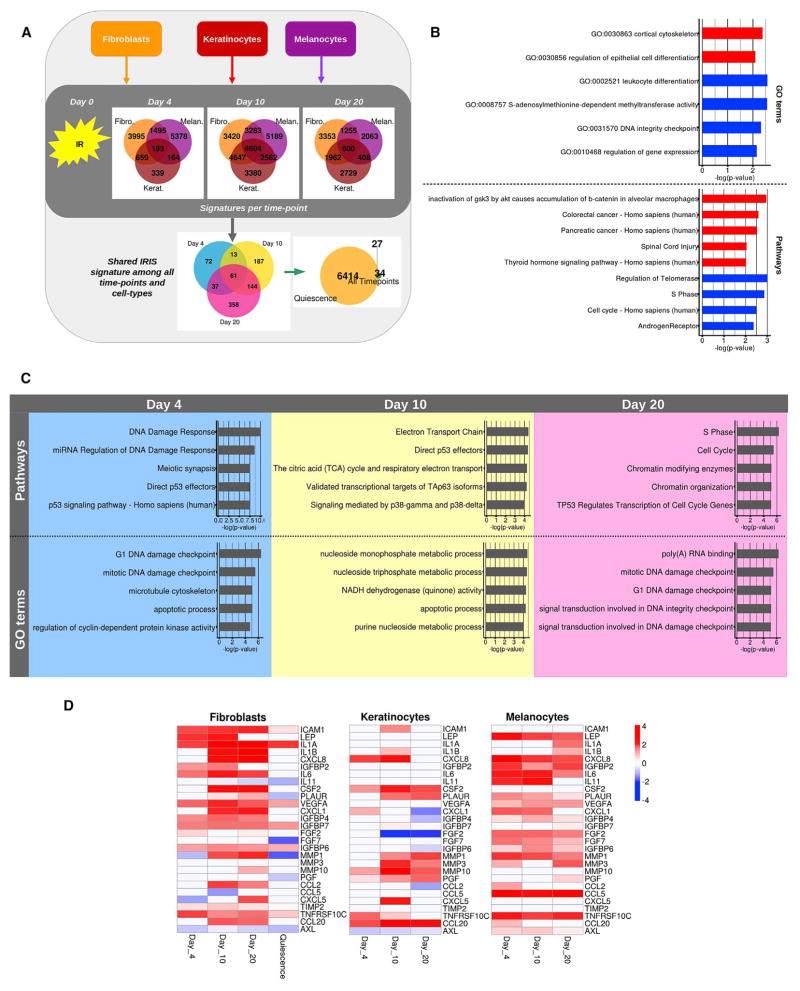
Figure 3. Temporal Dynamics of the Senescence Transcriptome.
(A) Experimental design. Fibroblasts (HCA-2; yellow), melanocytes (red), and keratinocytes (magenta) were exposed to ionizing radiation (IR), and RNA was harvested 4, 10, or 20 days later. Transcriptomes of the different cell types and intervals after senescence induction were obtained by RNA-seq. A time-point signature with genes differentially expressed (p ≤ 0.01) in all three cell types and a shared IR-induced senescence (IRIS) signature with genes shared by all cell types and time points (p ≤ 0.01) were generated.
(B) GO terms and pathways enriched in the shared IRIS signature among all time points and cell types. The figure shows enriched GO terms in the upregulated (red) and downregulated (blue) genes of the signature. Bars indicate the logarithm base 10 of the p value.
(C) Top 5 GO terms and pathways enriched at each time point. The figure shows the enriched GO terms and pathways for days 4, 10, and 20. Bars indicate the logarithm base 10 of the p value.
(D) Heatmap showing the dynamics of genes encoding SASP factors for each cell type. Known SASP factors that were significantly differentially expressed during at least one time point in each cell type are shown. The heatmap shows the logarithm base 2 of the fold change for each time post-irradiation with respect to proliferating cells. Quiescence was measured only on fibroblasts. The violet arrows highlight MMP1, the only SASP factor commonly regulated at days 10 and 20 in all cell types.