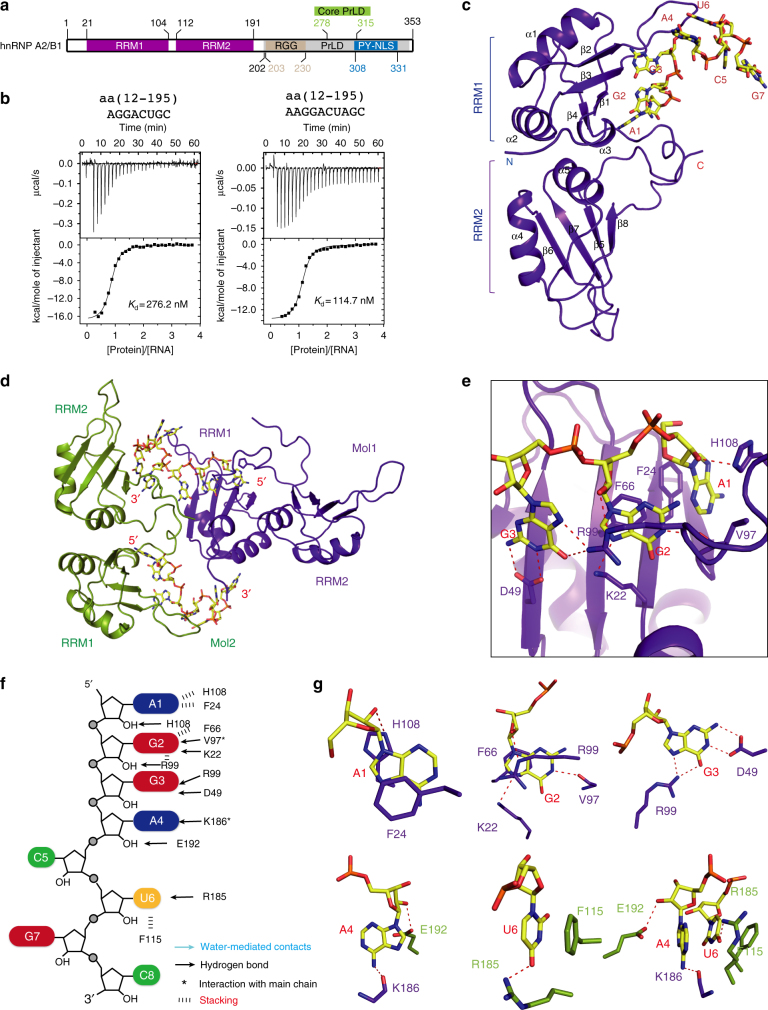
Fig. 1.
Overview of the structure and ITC of hnRNP A2/B1 in complex with 8mer RNA. a Schematic representation of the domain architecture of hnRNP A2/B1. RRM: RNA recognition motif, PrLD: prion-like domain, NLS: nuclear location signal, RGG: arginine-glycine-glycine box. b ITC results of hnRNP A2/B1(12–195) with 8mer and 10mer RNA targets. Solid lines indicate nonlinear least-squares fit the titration curve, with ΔH (binding enthalpy kcal mol−1), Ka (association constant), and N (number of binding sites per monomer) as variable parameters. Calculation values for Kd (dissociation constant) are indicated. c Cartoon representation of RRMs in complex with 8mer RNA. The RNA backbone is colored in yellow shown by stick. The RRMs are colored in purple-blue. d Molecules from two adjacent asymmetric units. The molecule from another asymmetric unit is colored in green. e Intermolecular contacts between RRM1 and 8mer RNA from A1 to G3. All dashed lines in this study indicate distance <3.2 Å. f Schematic showing RRMs interactions with 8mer RNA 5′-AGGACUGC-3′. g Close-up view showing the specific recognition of A1, G2, G3, A4, and U6