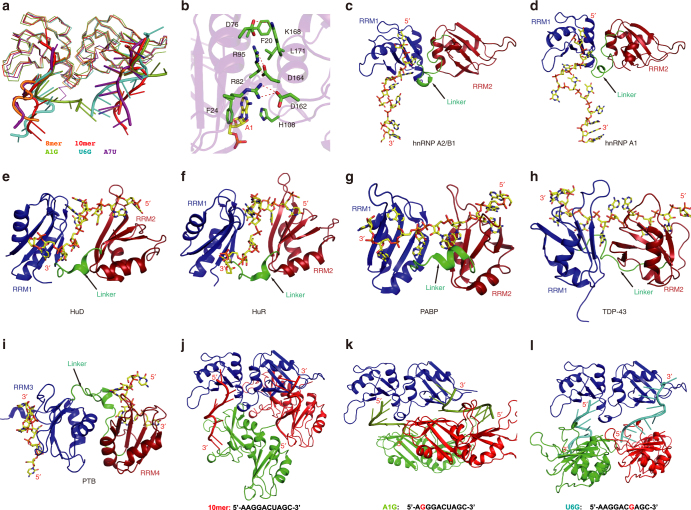
Fig. 5.
The RNA binding by RRMs adopts an antiparallel mode. a Superimposition of different structure complex. 8mer-RNA: 5′-AGGACUGC-3′ is colored in orange, 10mer-RNA: 5′-AAGGACUAGC-3′ is colored in red, A1G-RNA: 5′-AGGGACUAGC-3′ is colored in green, U6G: RNA 5′-AAGGACGAGC-3′ and A7U-RNA: 5′-AAGGACUUGC-3′ are colored in cyan and purple, respectively. b Interactions between RRM1 and RRM2; the amino acids participating in interactions are colored in green and RNA is colored in yellow. c The overall structure of hnRNP A2/B1-10mer RNA complex. d hnRNP A1 in complex with DNA. e–h The overall structure of HuD-RNA, HuR-RNA, PABP-RNA, and TDP-43-RNA. From c to h, RRM1 is colored in blue and RRM2 is colored in red, the linker is colored in green pointed out with a black arrow, the RNA backbone is colored in yellow shown by stick. i The overall structure of PTB–RNA complex with an antiparallel RNA-binding mode. RRM3 is colored in blue and RRM4 is colored in red, other labels are the same as from c to h. j–l Crystal packing interactions in 10mer, A1G and U6G. To illustrate the detailed packing interactions of hnRNP A2/B1 carrying two antiparallel RNA stands with other hnRNP A2/B1 molecules, three hnRNP A2/B1 molecules and two RNA strands of each complex are selected to show