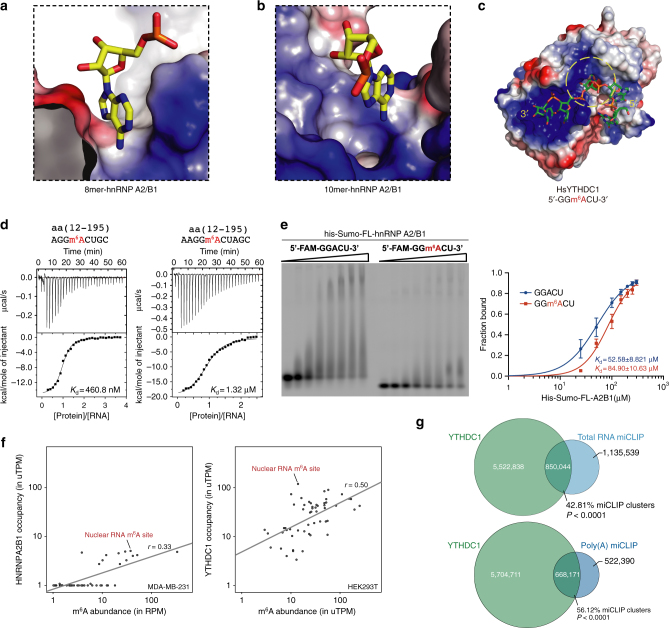
Fig. 6.
hnRNP A2/B1 does not specifically recognize m6A-modified RNA. a Surface representation of the environment around A4 in 8mer RNA complex. b Surface representation of the environment around A4 in 10mer RNA complex. c Surface representation of the canonical N6-methylated adenosine binding mode in HsYTHDC1. The aromatic cage is circled with yellow dashline. d ITC data of hnRNP A2/B1(12–195) with 8mer and 10mer RNA targets carried N6-methylated adenosine. e EMSA experiment shows the binding affinity of full-length hnRNP A2/B1 with 5′-FAM-labeled RNA substrates with or without m6A modification. Uncropped gel image is shown in Supplementary Fig. 7e. The data represent the mean of three independent experiments, with standard deviation (SD) values indicated by error bars. f YTHDC1 shows preferential binding to m6A sites in nuclear RNA compared to hnRNP A2/B1. For hnRNP A2/B1, the m6A-Seq reads that overlapped with each m6A site was plotted on the x-axis, and the HITS-CLIP reads that overlap with each site were plotted on the y-axis. A similar analysis was used to examine YTHDC1 binding at these m6A sites. miCLIP reads that overlapped with the m6A sites were plotted on the x-axis, and the YTHDC1 iCLIP reads that overlapped with the m6A sites were plotted on the y-axis. g YTHDC1-m6A tag cluster overlap. A Venn diagram indicating the cluster overlap is shown. Roughly, 43 and 56% of miCLIP tag clusters from total cellular RNA and poly(A) RNA showed a significant overlap with the YTHDC1 iCLIP clusters, respectively