Figure 1.
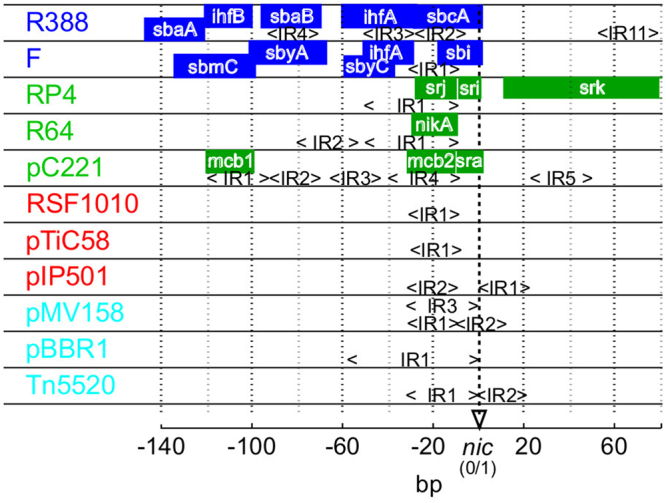
Schematic representation of available experimental data on oriT regions from four MOB groups. oriT data from MOB F (blue), P (green), Q (red) and V (cyan) supports that the conservation of structural properties within each MOB group is greater than between groups. Known binding sites for auxiliary proteins and relaxases are marked (colored squares) and are frequently characterized by inverted repeats (<IR>). Relaxase binding sites are nearest to the nic site (between 0 and 1 bp). General characteristics of MOB groups are: (F) system of multiple auxiliary proteins including the DNA-bending protein IHF44,55, (P) up to 5 proteins including relaxase involved in relaxation (RP4)47,54,61 (Q) a shorter oriT region of only 38 bp that covers besides relaxase 2 auxiliary proteins without clear binding sites (RSF1010)48,49,62, (V) no known auxiliary proteins50,51,63.
