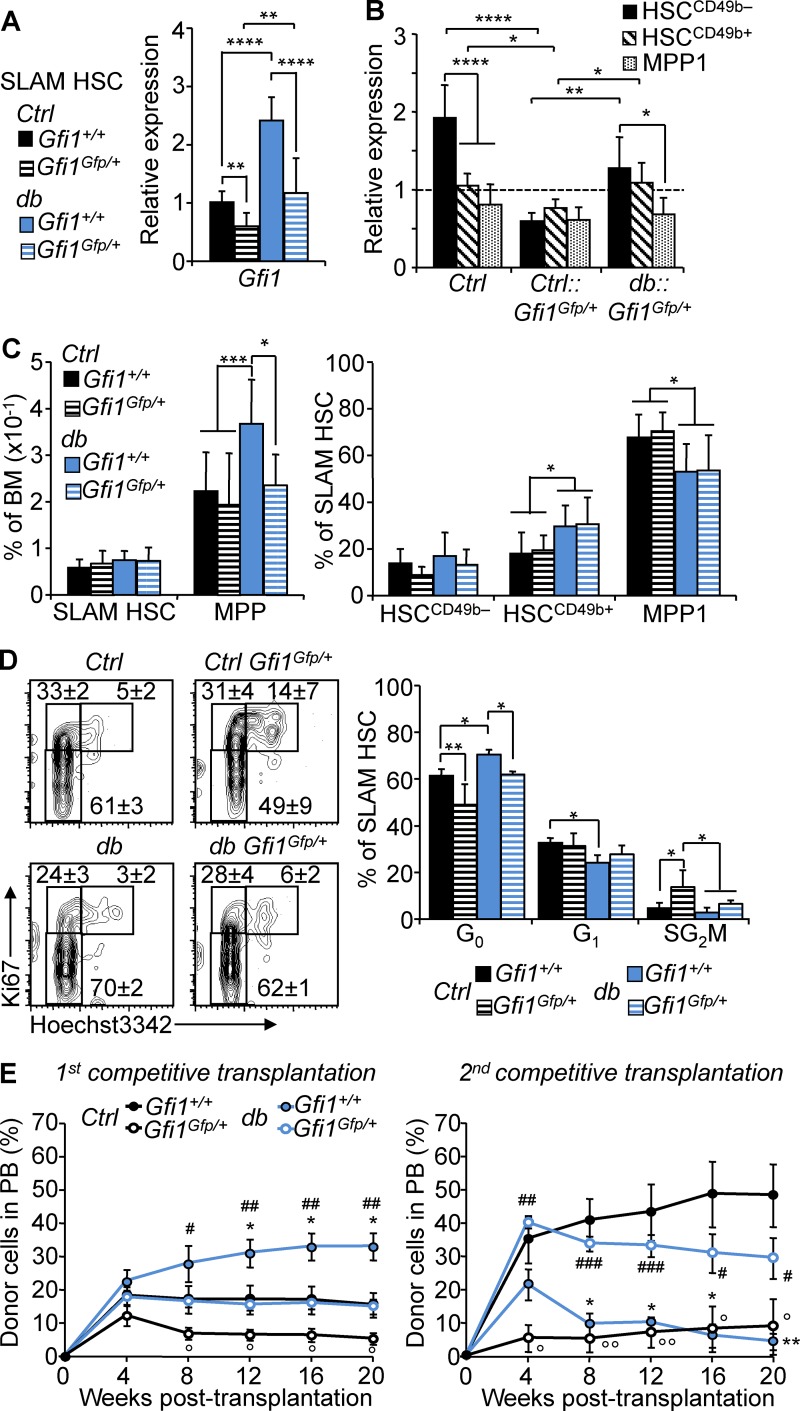
Figure 7.
Gfi1 controls HSC fate in obesity. (A) qRT-PCR analyses for Gfi1 gene expression in SLAM HSCs isolated from 4-mo-old db, Ctrl::Gfi1Gfp/+, and db::Gfi1Gfp+ mice. Results are expressed as fold change ± SD relative to total Ctrl SLAM HSCs. n = 6–12 pools of 100 cells. Student’s t test; **, P ≤ 0.01; ****, P ≤ 0.0005. Three independent experiments. (B) qRT-PCR analyses for Gfi1 gene expression in HSC subsets isolated from 4-mo-old Ctrl, Ctrl::Gfi1Gfp/+, and db::Gfi1Gfp/+ mice. Results are expressed as fold change ± SD relative to total Ctrl SLAM HSCs. n = 6–12 pools of 100 cells. Two-way ANOVA with Tukey’s post hoc test; *, P ≤ 0.05; **, P ≤ 0.005; ****, P ≤ 0.0001. Two independent experiments. (C) Mean percentages ± SD of SLAM HSC/MPPs (left) and HSC subsets (right) in the BM of 4-mo-old Ctrl, db, Ctrl::Gfi1Gfp/+, and db::Gfi1Gfp+ mice. n = 8–18. (D) Representative FACS plots (left) and mean percentages ± SD (right) showing SLAM HSC distribution in cell cycle phases. n = 4–6 mice/group. One-way ANOVA with Tukey’s post hoc test; *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.001. Two independent experiments. (E) PB chimerism in primary (left) and secondary (right) competitive reconstitution assays using SLAM HSCs isolated from Gfi1Gfp/+ and db compound mice. Results are expressed as means ± SEM. n = 4–8/group. Student’s t test; Ctrl vs. db: *, P ≤ 0.05; **, P ≤ 0.005; Ctrl vs. Ctrl::Gfi1Gfp/+: °, P ≤ 0.05; °°, P ≤ 0.05; db vs. db::Gfi1Gfp/+: #, P ≤ 0.05; ##, P ≤ 0.005; ###, P ≤ 0.0005. Two independent experiments.