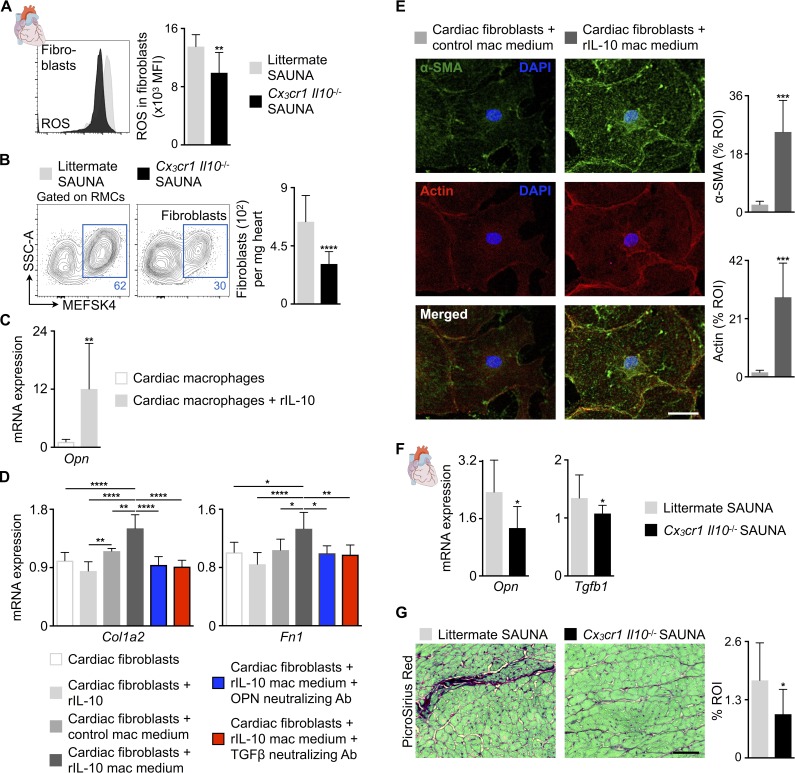
Figure 9.
IL-10 produced by cardiac macrophages indirectly activates fibroblasts. (A) Mean fluorescence intensities (MFI) indicating ROS production in cardiac fibroblasts from littermate and Cx3cr1 Il10−/− SAUNA-exposed mice. Data are pooled from two independent experiments (n = 11 mice per group). (B) Flow cytometric quantification of fibroblasts in hearts from littermate and Cx3cr1 Il10−/− SAUNA-exposed mice. Left: Representative flow cytometry plots; right: number of fibroblasts per milligram of heart tissue. Data are pooled from two independent experiments (n = 11–15 mice per group). RMCs, resident mesenchymal cells. (C) Relative Opn expression levels by qPCR in control and rIL-10–exposed FACS-purified cardiac macrophages. Data are pooled from two independent experiments (n = 10 per group). (D) Relative Col1a2 and Fn1 expression levels by qPCR in FACS-purified cardiac fibroblasts incubated with rIL-10, control macrophage (mac) medium or rIL-10–exposed mac medium with and without OPN or TGFβ neutralizing antibody (Ab). Data are pooled from two independent experiments (n = 4–7 per group). (E) Left: Representative immunofluorescence images of FACS-purified cardiac fibroblasts incubated with control or rIL-10–exposed mac medium, and stained with α-SMA (green), Phalloidin to identify actin filaments (red), and DAPI (blue); right: bar graphs show percentage of positive α-SMA or actin staining per ROI. Data are pooled from two independent experiments (n = 7 per group). Bar, 50 µm. (F) Relative Opn and Tgfb1 expression levels by qPCR in hearts from littermate and Cx3cr1 Il10−/− SAUNA-exposed mice. Data are pooled from at least two independent experiments (n = 8–15 mice per group). (G) Histological analysis of collagen deposition (PicroSirius Red) in hearts from littermate and Cx3cr1 Il10−/− SAUNA-exposed mice. Left: Representative images; right: bar graph shows percentage of positive staining per ROI. Data are pooled from two independent experiments (n = 6 mice per group). Bar, 50 µm. Results are shown as mean ± SD. For statistical analysis, a two-tailed unpaired t test was performed to compare two groups, and one-way ANOVA followed by Tukey’s test was performed for multiple comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.