Figure 3.
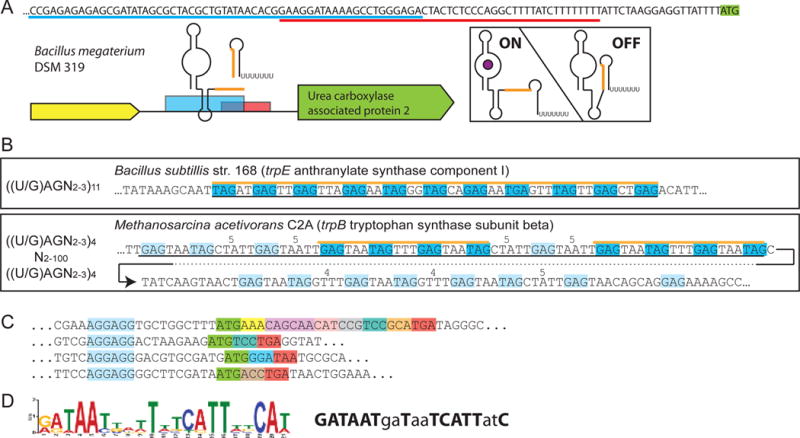
Results from RiboGap queries (A) The expression platform of a ykkC-yxkD (guanidine-I) riboswitch is observed from the overlap of the Rho-independent terminator (sequence underlined in red) and aptamer (partial sequence, underlined in blue). Genetic context, sequence overlap of structures is shown in orange. Hypothetical ON and OFF states are pictured on the right. (B) Examples of TRAP binding motifs found with the patterns used for the search on the left. Top is the canonical pattern, with a hit corresponding to a confirmed TRAP-regulatory site. Bottom, the pattern used for the search was less stringent and revealed a putative archaeon TRAP-like binding site. Black lines under the sequences indicate the putative full TRAP motif. Blue boxes and orange line correspond to binding sites found by the search, pale blue to other potential binding sites. Spacers larger than the typical 2-3 bases are indicated above the sequence. (C) Examples of mini ORFs found by the RiboGap query. Mini-ORF Shine-Dalgarno boxed in blue, start codon in green, stop codon in red and other codons in different colors. (D) Conserved “iron-box” found by MEME and drawn by sequence logo [59] on the left, Fur-box consensus on the right. Positions matching the “iron-box” consensus are in bold.
