Extended Data Figure 3. Transcriptional differences between Epiblast and E×E are directed in part through DNA methylation.
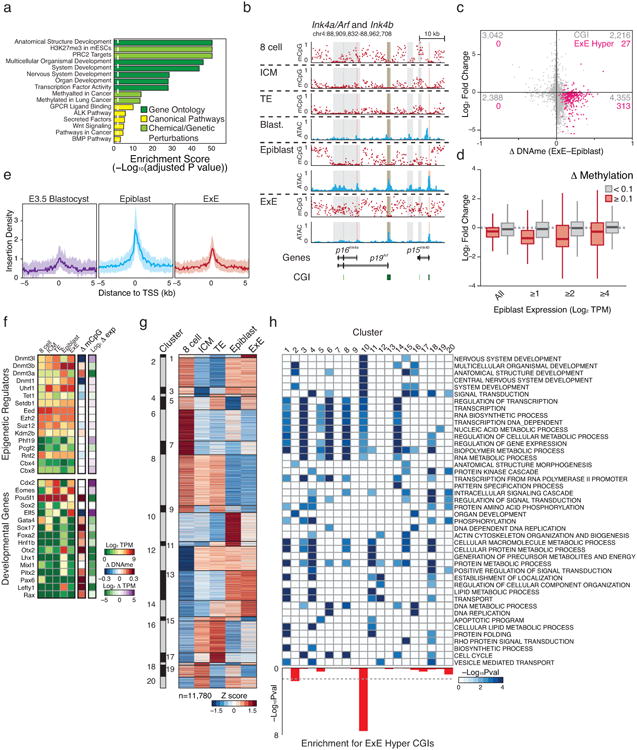
a. Select gene set enrichment analysis of E×E-hypermethylated transcription start sites including Gene Ontology, Canonical Pathways, and Genetic and Chemical Perturbations. E×E-hypermethylated promoters are highly enriched for transcription factors and signaling pathways involved in patterning the early embryo. Moreover, these CGI regulated genes are canonical targets of PRC2, which coordinates selective expression of key developmental regulators during gastrulation.
b. DNA methylation and open chromatin dynamics for the tumor suppressors p16Ink4a, p19Arf, and p15Ink4b. While these loci are either basally or non-transcribed during early development, three regions are dynamically methylated in E×E (highlighted in gray), including a >10 kb region that encompasses the entirety of the p16Ink4a locus and is either wholly unmethylated in Epiblast or extensively methylated in E×E. CpG islands are highlighted in green, and the positions of included TSSs are highlighted in red.
c. Scatterplot of Log2 expression dynamics versus differential CGI methylation between Epiblast and E×E. While most dynamically methylated CGI-promoter containing genes have functions in later embryonic development and are not yet highly expressed, de novo methylation in E×E is generally associated with transcriptional repression. E×E hyper CGIs are highlighted in pink. Promoter CGIs are assigned to the most proximal gene within a + or – 2 kb boundary.
d. Boxplots demonstrating the relationship between promoter methylation and expression in the restriction of extraembryonic and embryonic compartments. Promoters are defined as + or – 1 kb of an annotated TSS and scored as dynamically methylated in E×E if the difference with Epiblast is ≥0.1. Expression changes between dynamically methylated and background promoter sets are provided over increasing thresholds according to their expression in Epiblast. While many CGI promoters are not dynamically expressed in either Epiblast or E×E and are associated with genes that have downstream developmental functions, transcriptional repression is a consistent feature of promoter methylation, even at this low threshold.
e. Median open chromatin signal as measured by ATAC-seq for E×E Hyper CGI-associated TSSs in the transition from pre- to postimplantation. E×E Hyper CGI-associated genes are heavily enriched for roles in patterning the embryo proper and are primarily not expressed until the onset of gastrulation. In the transition from Blastocyst to Epiblast, these promoters gain open chromatin signal, suggesting transcriptional priming or activation, which is not observed within the E×E, where they are de novo methylated. Shaded area reflects the 25th and 75th percentile per fixed 100 bp bin.
f. Expression and differential promoter methylation of key epigenetic regulators and master regulators over early embryonic and extraembryonic development. Most epigenetic regulators exhibit minimal expression differences between Epiblast and E×E, with the Dnmts being notable exceptions. Key isoforms of Dnmt3a and Dnmt3b are upregulated in Epiblast in conjunction with global remethylation, while the suppression of Dnmt3a in E×E corresponds with de novo promoter methylation. Alternatively, the maintenance methyltransferase Dnmt1 and the non-catalytic cofactor Dnmt3l are induced within the blastocyst and maintained at higher levels in E×E, with reciprocal methylation of the Dnmt3l promoter in Epiblast. The histone 3 lysine 36 demethylase Kdm2b displays differential expression of catalytically active and inactive isoforms within Epiblast and E×E, respectively, with isoform switching seemingly imposed by de novo methylation around the somatically utilized CGI promoter. The E×E is characterized by persistent expression of the master regulators Cdx2, Eomes, and Elf5 (Refs 47-50), while the still pluripotent epiblast remains Pou5f1 (Oct4) positive. Many additional regulators of subsequent developmental stages are basally expressed within the Epiblast and their promoters de novo methylated in E×E. The difference in promoter methylation refers to the annotated TSS that exhibits the greatest absolute difference between E×E and Epiblast. TPM: Transcripts Per Million. Additional high-resolution genome-browser tracks are displayed for select transcriptional and epigenetic regulators in Extended Data Figure 4 and 7, respectively.
g. Unsupervised hierarchical clustering of 11,780 genes over late preimplantation and early post-implantation development, partitioned into 20 distinct dynamics (“clusters”). Cluster 10 includes genes that are specifically induced within the Epiblast but not the E×E. Heatmap intensity reflects the row-normalized Z score.
h. Significant Gene Ontology enrichment for the 20 gene expression dynamics characterized in e, including for those regulated by E×E-methylated CGI promoters, as calculated using the binomial test. Cluster 10 is enriched for both developmental functions and E×E promoter methylation.
