Extended Data Figure 6. Generation of dual expression and methylation libraries from outgrowth and embryonic knockout data.
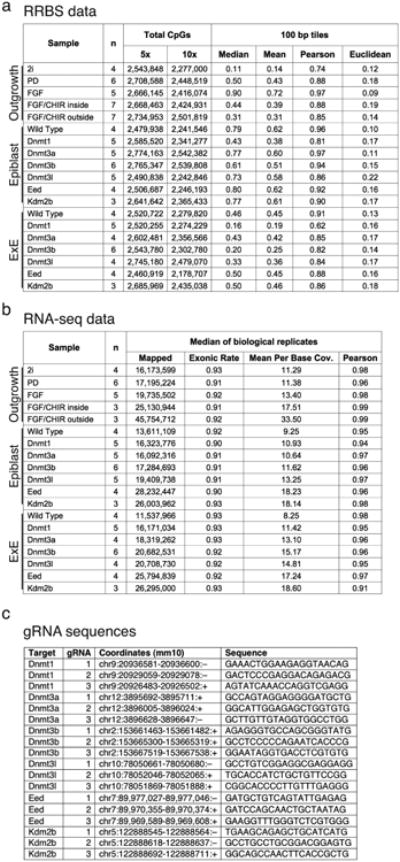
a and b. Sequencing metrics and coverage information for dual RRBS and RNA-seq libraries generated for the evaluation of ICM outgrowths and CRISPR/Cas9 disrupted E6.5 embryos, including similarity metrics between replicates (Euclidean distance and Pearson Correlation for RRBS and Pearson correlation for RNA-seq). Mean and median methylation of 100 bp tiles is also included for the RRBS samples.
c. CRISPR/Cas9 disrupted embryos were generated by zygotic injection of three single guide RNA (gRNA) sequences specific to early exons that are shared across different isoforms. The genomic coordinates and protospacer sequences are provided (see Methods).
