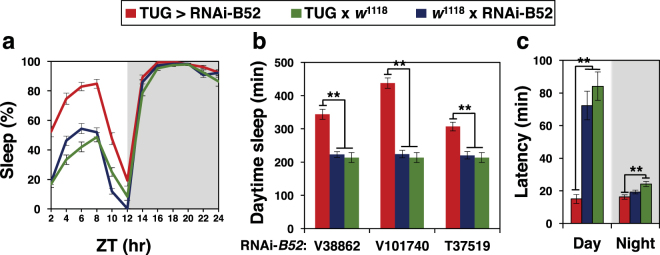
Figure 3.
Knock down of B52 in clock cells preferentially modulates daytime sleep during a daily light-dark cycle. (a–c) Young male progeny from the indicated cross (top of panels) were kept at 18 °C and entrained for 5 days in 12 hr:12 hr light/dark (LD) cycles [where Zeitgeber time (ZT) 0 is lights-on] followed by several days in constant darkness (DD; see Fig. 4). Daily wake sleep levels (a), total daytime sleep (b) and sleep latency (c) were measured, and average values during the last 3 days of LD are shown. For each genotype, data from 16 individual flies was used. Daily sleep levels (a) and sleep latency (c) for TUG > RNAi-B52(V101740) and control crosses are shown. Similar results were obtained with other RNAi-B52 lines (data not shown). (b) Total daytime sleep levels (ZT0–12) for several independent lines of RNAi-B52 (V38862, V101740, T37519) and appropriate control crosses. The following p values were determined: [Panel (a), ANOVA, TUG > V101740 compared to both controls; for daytime values from ZT0-12, 3.4 × 10−6; for nighttime values from ZT12-24, 0.057]. [Panel (b), two-tailed t-test; TUG > V38862 vs V38862 x w1118, 5.0 × 10−10; TUG > V38862 vs TUG x w1118, 1.2 × 10−8; TUG > V101740 vs V101740 x w1118, 1.68 × 10−20; TUG > V101740 vs TUG x w1118, 1.96 × 10−18; TUG > T37519 vs T37519 x w1118, 2.19 × 10−6; TUG > T37519 vs TUG x w1118, 1.06 × 10−8]. [Panel (c), two-tailed t-test; for day values, TUG > V101740 vs V101740 x w1118, 9.56 × 10−8; TUG > V101740 vs TUG x w1118, 4.8 × 10−10; for night values, TUG > V101740 vs V101740 x w1118, 0.081; TUG > V101740 vs TUG x w1118, 0.00025]. (b,c) Values for TUG > RNAi-B52 were significantly different compared to one or more control crosses; *p < 0.05; **p < 0.01; two-tailed t-test. The corresponding daily activity profile for panel (a) is shown in Figure S2a.