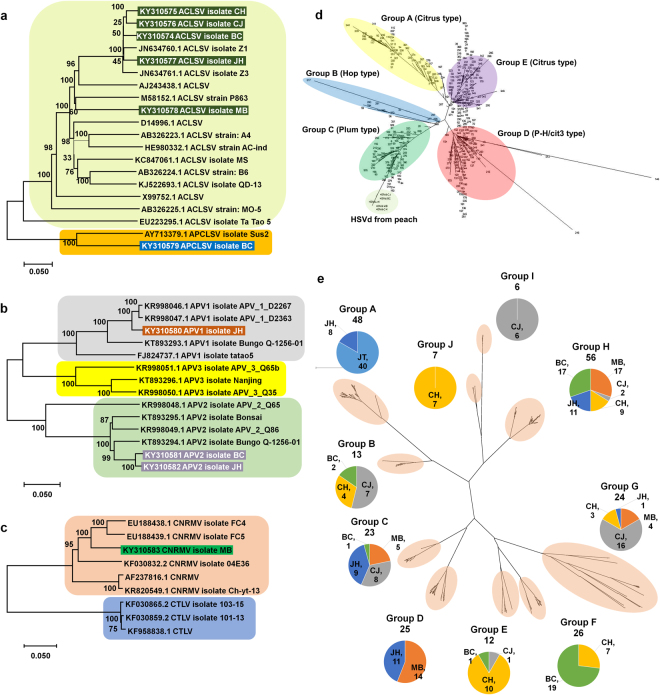
Figure 5.
Phylogenetic analyses of identified viruses and viroids. (a) Phylogenetic relationships for five ACLSV isolates: CH, CJ, BC, JH, and MB, as well as APCLSV isolate BC in this study with known ACLSV and APCLSV genomes. (b) Phylogenetic relationships of APV1 isolate JH, APV2 isolate BC, and APV2 isolate JH with known APV1, APV2, and APV3 genomes. (c) The phylogenetic relationships of CNRMV isolate MB and known CNRMV and CTLV genomes. (d) The phylogenetic relationships of five HSVd isolates in this study with all HSVd genomes. The accession numbers for five HSVd genomes are KX371905–KX371909. (e) The phylogenetic relationships of 240 PLMVd genomes obtained via RT-PCR following by Sanger Sequencing in this study. The accession numbers of 240 PLMVd genomes are PLMVd-BC (KY355162–KY355201), PLMVd-CH (KY355202–KY355241), PLMVd-CJ (KT033033–KT033052 and KY355242–KY355261), PLMVd-JH (KY355262–KY355301), PLMVd-JT (KY355302–KY355341), and PLMVd-MB (KY355342–KY355381). The pie chart indicates the composition of identified PLMVd genome from each peach cultivar with the respective number of PLMVd genomes in each identified group.