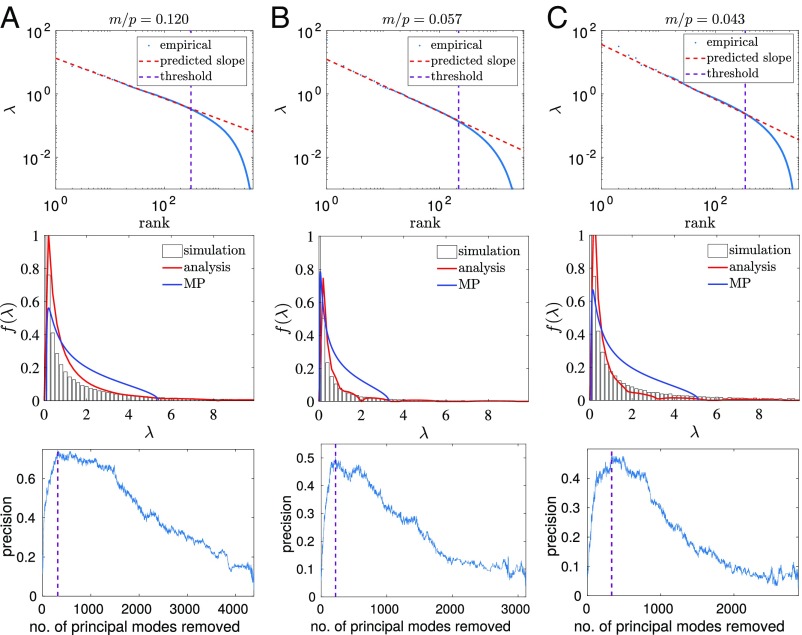
Fig. 6.
Protein sequence alignments follow the power law, and moreover spectral deviation from the power law can be used to deconvolve the influence of phylogeny from the covariance matrix, and facilitate contact prediction. A–C show analysis of protein sequence data from (A) Trypsin, (B) DHFR, and (C) TRML-HAEIN, a knotted tRNA-methyltransferase. In A–C, Top we show that the eigenvalues of each protein sequence alignment follow a power law. The purple dashed line indicates the point at which the spectrum deviates from this power law, indicating a threshold above which phylogeny dominates the spectrum. The parameter is inferred from this power law using the equation , where and is given by Eq. 1. The inferred values of are used to plot the red lines in A–C, Middle, which provide a good fit to the empirical spectral distributions. A–C, Bottom show that the phylogenetic threshold, derived from A–C, Top, provides an excellent indication of which modes should be removed from the covariance matrix to deconvolve the influence of phylogeny and dramatically improve contact prediction using just the covariance matrix.