Significance
The production of mucus helps to trap pathogens, preventing their entry into the body, while it also acts as an interface for many important physiological events (e.g., gas and nutrient exchange). In mammalian models, a detailed study of mucus and its component parts is hindered by the difficulty in accessing these internally located tissues. The Xenopus tropicalis tadpole skin offers a complementary nonmammalian model system to study mucosal epithelia. Using this, we identify a mucin, similar to human mucins, that protects against infection. This system offers an experimentally tractable approach to study mucins and the mucus barrier and their role in conferring protection at mucosal surfaces.
Keywords: mucus, mucin, Xenopus tropicalis, Aeromonas hydrophila, innate defense
Abstract
Mucosal surfaces represent critical routes for entry and exit of pathogens. As such, animals have evolved strategies to combat infection at these sites, in particular the production of mucus to prevent attachment and to promote subsequent movement of the mucus/microbe away from the underlying epithelial surface. Using biochemical, biophysical, and infection studies, we have investigated the host protective properties of the skin mucus barrier of the Xenopus tropicalis tadpole. Specifically, we have characterized the major structural component of the barrier and shown that it is a mucin glycoprotein (Otogelin-like or Otogl) with similar sequence, domain organization, and structural properties to human gel-forming mucins. This mucin forms the structural basis of a surface barrier (∼6 μm thick), which is depleted through knockdown of Otogl. Crucially, Otogl knockdown leads to susceptibility to infection by the opportunistic pathogen Aeromonas hydrophila. To more accurately reflect its structure, tissue localization, and function, we have renamed Otogl as Xenopus Skin Mucin, or MucXS. Our findings characterize an accessible and tractable model system to define mucus barrier function and host–microbe interactions.
Mucosal and mucociliary epithelia (hereafter mucosal) are major sites of pathogen interaction in animals. These surfaces are directly exposed to the environment with mucus representing the first barrier that they must overcome to access the epithelial layer. Despite this critical host-protective function, research into the interactions of microbes with mucus has been a neglected area of research.
Mucus is composed of hundreds of proteins, but the major structural components are the polymeric, high-molecular-weight (2–100 MDa), O-linked glycoproteins called mucins (1). Mucins form networks with viscoelastic properties that entrap particles and pathogens while also acting as a framework for interaction with other proteins (2, 3).
It is important to study the fundamental biology of mucosal epithelia because of their crucial role in defense against infection, but also because they are dysfunctional in conditions such as cystic fibrosis and ulcerative colitis (4, 5). However, the anatomical location of mammalian mucosal surfaces makes studying them in situ challenging, requiring invasive techniques to access them and creating difficulties in reconstituting them in vitro. Indeed, this is a major reason why the interactions of microbes with mucus in their native environment have been understudied. To address this issue, we have investigated the skin of the developing tadpole, Xenopus tropicalis, which has an exposed, accessible mucosal epithelium (6, 7).
X. tropicalis embryos develop a host-protective epidermis after gastrulation, and this remains during tadpole stages until metamorphosis (7). The tissue architecture of the tadpole epidermis is highly similar to mammalian respiratory mucosal epithelia (8). Motile, multiciliated cells beat in a polarized direction to generate fluid flow from head to tail, while two populations of secretory cells, goblet cells, and small secretory cells (SSCs) have been identified (7, 9, 10). These cells are hypothesized to produce a mucus-like protective layer, which is likely moved away by the actions of the beating cilia (11). Previously, we showed that SSCs develop alongside goblet cells and multiciliated cells in early embryonic development, and when SSCs are depleted, the embryos succumb to infection (7). We identified a number of proteins secreted from the epidermis, including an abundant protein called Otogelin-like (Otogl), named because of its sequence similarity with a glycoprotein called Otogelin found in the acellular membranes of the human inner ear (12). At the time, the functional role of Otogl in the tadpole skin was unknown (7).
In the current study, we demonstrate that Otogl is a type of gel-forming mucin that forms high-molecular-weight complexes and is glycosylated through mucin-type O-glycosylation. We also demonstrate that this mucin forms a physical barrier at the surface of the tadpole skin and that this barrier traps bacteria and prevents infection when challenged with a relevant opportunistic pathogen.
Results
Otogl Has Similar Sequence and Domain Organization to Gel-Forming Mucins.
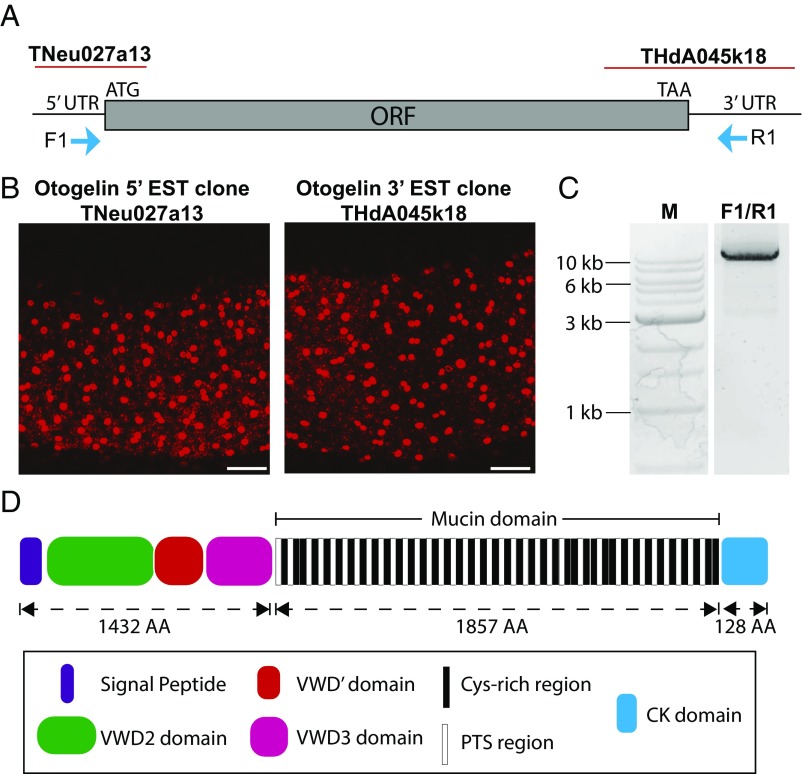
The initial annotation of the X. tropicalis genome had several gaps in the center of the otogl gene (7, 11), which made it unclear whether it was one gene or two. To address this issue, we made RNA in situ hybridization probes targeting both the putative 5′ UTR/start site and the 3′ UTR/termination site (Fig. 1 A and B). Both probes gave a similar expression pattern by fluorescent in situ hybridization, with abundant staining in SSCs and weaker staining in goblet cells, as shown previously for a 3′ otogl probe (7). This suggested that otogl is one long transcript, and we confirmed this by reverse transcriptase long-range PCR (Fig. 1C) using primers in the 5′ and 3′ UTRs (F1/R1, Fig. 1A). The dominant PCR product was sequenced by long-read single-molecule real time (SMRT) sequencing and gave an ORF of 10,254 bases. This translates to a protein of 3,417 amino acids (∼370 kDa).
Fig. 1.
Otogl is a large protein with mucin-like domains. (A) Model of Otogl transcript showing sites corresponding to EST clones TNeu027a13 and THdA045k18, as well as 5′ and 3′ UTRs, start (ATG) and stop (TAA) codons, and primer sites (F1 and R1). (B) RNA in situ hybridization expression patterns of TNeu027a13 and THdA045k18. (Scale bar: 100 μm.) (C) PCR using F1-R1 primers on X. tropicalis cDNA. “M” is marker DNA ladder. (D) Predicted domains of Otogl. VWD is von Willebrand factor type D domain. The mucin domain shows the 39 Cys-rich and 34 PTS-rich subdomains. CK, cysteine knot domain. The size of the domains in numbers of amino acids (AA) is shown.
Analysis of the translated amino acid sequence revealed a large, multidomain protein with high similarity to gel-forming mucins (Fig. 1D and Fig. S1). Following a signal sequence (indicating secretion), Otogl has three von Willebrand factor D (VWD) domains, arranged as VWD2-VWD′-VWD3 (Fig. 1D), which is typical for mucins. The VWD domains have conserved cysteine residues with human gel-forming mucins (Fig. S2), and these are essential for intra- and intermolecular disulfide bonding that are important in secondary structure and multimerization (1, 13).
The central (mucin) domain (Fig. 1D) makes up over 50% of Otogl and is highly repetitive. In gel-forming mucins, these domains are also highly repetitive and are enriched in proline, threonine, and serine (PTS) residues (PTS domains, sites of extensive O-glycosylation) with occasional interruption by Cys-rich subdomains (CysD domains) (14). Analysis of the central domain of Otogl (Fig. 1D) revealed the presence of 39 well-conserved repeats of a Cys-rich sequence and 34 PTS repeats (Fig. S3). The PTS repeats likely represent the sites of mucin-type O-glycosylation. At the C terminus of Otogl is a cysteine knot (CK) domain, with cysteine residues precisely conserved in position with those of human gel-forming mucins (Fig. S2). CK domains are essential for dimerization of gel-forming mucins (15).
The identity of the encoded Otogl glycoprotein was confirmed by mass spectrometry (MS) of tadpole skin secretions. In total, 16.5% (563/3,417) of the protein was covered by the MS analysis with peptides identified throughout the protein (Fig. S1). In the central mucin domain, the vast majority of the peptides identified (98.4%, 61/62 peptides) were located in the Cys-rich regions. Only a single peptide was identified in the PTS-rich regions, which indicates that these regions are inaccessible to trypsin digestion due to O-glycosylation, as reported with other mucins (16). Overall, these analyses indicated that Otogl would have the properties of a gel-forming mucin.
Otogl Is an O-Linked Glycoprotein with Structural Similarities to Mucins.
Gel-forming mucins are defined as large, heavily O-glycosylated, disulfide-linked polymeric proteins that have an extended conformation in solution (17). These are the criteria that we used to assess the properties of Otogl.
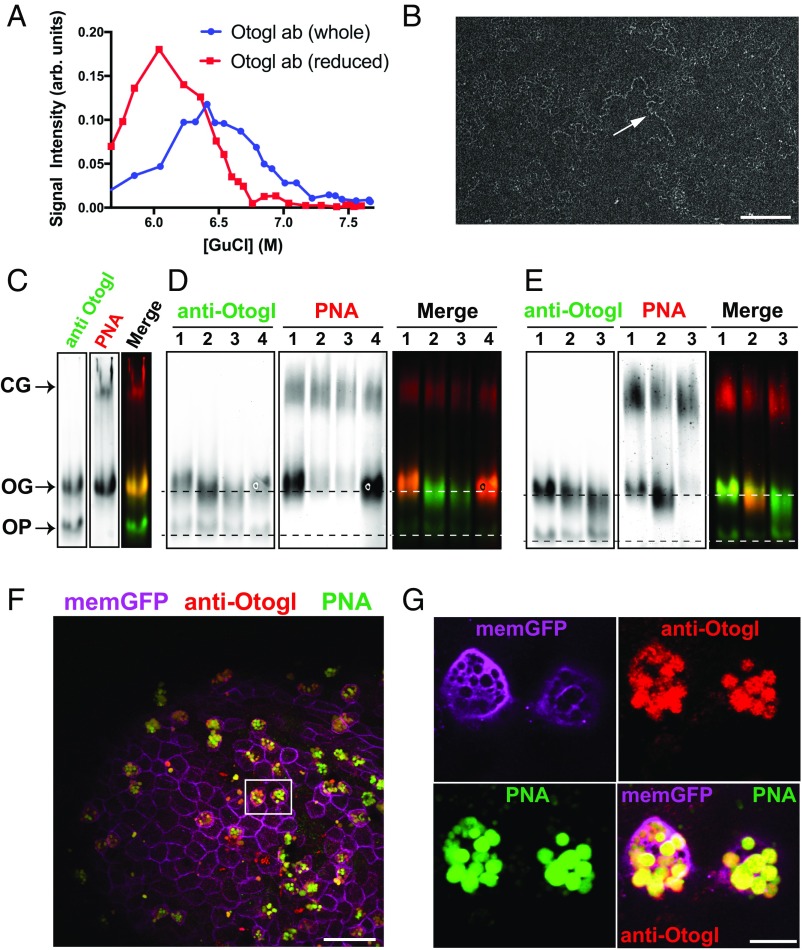
To demonstrate the effect of disruption of disulfide bonds on the size/shape of Otogl, we performed sedimentation analysis on tadpole secretions. Analysis of fractions from the guanidinium chloride (GdmCl) density gradient probed with an Otogl-specific antibody showed that the distribution of anti-Otogl reactive material shifted from higher density (multimers) to lower density (monomers) after the sample was reduced (Fig. 2A). This behavior is typical for gel-forming mucins due to their depolymerization by disruption of intermolecular disulfide bonds (1, 17). We purified Otogl from tadpole secretions by CsCl density gradient centrifugation, analyzed its structure by transmission electron microscopy (TEM), and determined its molecular weight by size exclusion chromatography coupled with multiangle laser light scattering (SEC-MALLS). In TEM, extended chain-like networks (Fig. 2B), which resembled structures observed for human gel-forming mucins, were visible (18). By SEC-MALLS, Otogl was found to have a molecular weight in the range of 0.6–4 MDa, with an average of 1.6 MDa. Taken together, these data suggest that Otogl is a polymeric macromolecule. To test that it was an O-linked glycoprotein, we performed lectin blots and glycosidase digests (Fig. 2 C–E).
Fig. 2.
Otogl is a multimeric O-glycosylated glycoprotein. (A) Rate zonal centrifugation of nonreduced and reduced tadpole skin secretions probed with anti-Otogl antibody. (B) TEM of CsCl density gradient purified Otogl shows long-chain–like networks (arrow). (Scale bar: 200 nm.) (C) Coprobing of a Western blot of tadpole lysate with anti-Otogl antibody and PNA; a merged image is also shown. CG, cement gland mucin; OG, glycosylated form of Otogl; OP, precursor form of Otogl. (D) Treatment of tadpole lysate with O-glycosidase (2 h, lane 2, and 4 h, lane 3) and PNGase F (lane 4) compared with control (lane 1), probed with anti-Otogl and PNA; a merged image is also shown. Dashed lines represent the position of bands for control OG and OP. (E) Treatment of tadpole lysate with sialidase alone (lane 2) and sialidase + O-glycosidase (lane 3) compared with control (lane 1); a merged image is also shown. Dashed lines represent the position of bands for control OG and OP. In D and E, the signal for PNA from the cement gland mucin shows the approximately equivalent loading between lanes. (F) Image of section from fixed whole-mount tadpole skin with immunofluorescence for anti-Otogl and anti-GFP [labeling membrane-GFP (memGFP) to identify membranes] together with lectin histochemistry (PNA) shows colocalization of Otogl and PNA staining. The boxed area highlights two adjacent SSCs. (Scale bar: 50 μm.) (G) Zoomed-in image of boxed area from F shows Otogl and PNA colocalize within the vesicles of SSCs. ab, antibody. (Scale bar: 10 μm.)
We have shown previously that high-molecular-weight macromolecules in tadpole secretions are recognized by lectin, peanut agglutinin, or peanut agglutinin (PNA) (7). PNA binds to the glycan epitope galactosyl (β-1,3) N-acetylgalactosamine [Gal(β-1,3)-GalNAc], which is commonly associated with O-glycosylated proteins. In tadpole secretions, PNA appears to recognize a mucin secreted from the cement gland, as well as a glycoprotein secreted from the tadpole skin, which was predicted to be Otogl. To investigate this, we performed a dual PNA blot and immunoblot for Otogl on reduced tadpole lysate after agarose gel electrophoresis (Fig. 2C). PNA identified two species: one that migrated slowly in the gel and corresponded to the cement gland (CG) mucin and a faster-migrating species that was also recognized with the Otogl antibody. The faster-migrating band likely represents the glycosylated form of Otogl (OG). The Otogl antibody also recognized another band migrating even faster than OG that most likely represents the non–O-glycosylated precursor of Otogl (OP).
To further investigate the nature of Otogl glycosylation, we conducted O-glycosidase and N-glycosidase digests on reduced tadpole lysate. Compared with controls (lane 1, Fig. 2D), treatment with O-glycosidase (lanes 2 and 3, Fig. 2D) caused a significant reduction in PNA binding and increased electrophoretic mobility of OG-Otogl (Fig. 2D). This indicates that Otogl is O-glycosylated with the disaccharide Gal(β-1,3)-GalNAc. Mucins are also sialylated, which can mask the Gal(β-1,3)-GalNAc disaccharide. To test this, we investigated the effect of sialidase treatment alone and in combination with O-glycosidase on reduced tadpole lysate compared with controls. Sialidase treatment alone (lane 2, Fig. 2E) caused increased migration of OG-Otogl and showed an increased signal for PNA compared with controls (lane 1, Fig. 2E), which is likely due to the exposure of Gal(β-1,3)-GalNAc epitopes. Subsequent treatment with O-glycosidase (lane 3, Fig. 2E) dramatically reduced PNA binding to barely detectable levels and increased the electrophoretic mobility of OG-Otogl. These data indicate that Otogl is O-glycosylated and has both Gal(β-1,3)-GalNAc and sialic-acid–containing glycan structures. The observation that, after treatment with sialidase and O-glycosidase, OG-Otogl could still be differentiated from its precursor (OP) by electrophoresis indicated that Otogl is likely to be glycosylated with additional glycans. To test for the presence of N-glycans, PNGase F was applied to the reduced tadpole lysate. As expected for a mucin, PNGase F did not cause a loss in signal for PNA, but there is evidence for a slight increase in electrophoretic mobility of the OG-Otogl (less than after O-glycosidase treatment) and OP-Otogl (lane 4, Fig. 2D). Taken together, these data indicate that Otogl, like conventional gel-forming mucins, is mainly O-glycosylated, with a potential smaller contribution from N-glycans.
To show that Otogl and PNA staining colocalize in the vesicles of the secretory cells in the tadpole skin, we performed dual lectin- and immunofluorescence on fixed tadpoles. Colocalization of anti-Otogl and PNA staining was evident in SSCs (Fig. 2F) and, more specifically, in the vesicles of these cells (Fig. 2G), further evidence that Otogl is glycosylated in its final form. Having established that Otogl has both domain similarities and structural similarities with mammalian gel-forming mucins, we next investigated the barrier function of Otogl on the skin surface.
Otogl Forms a Barrier at the Tadpole Skin Surface.
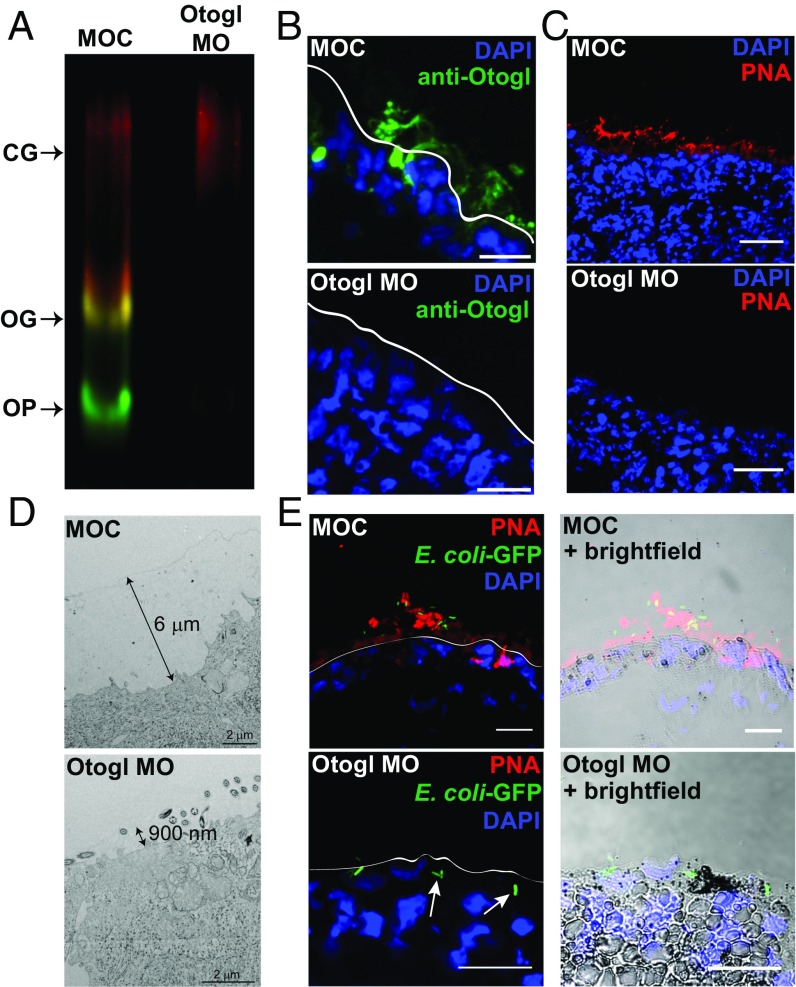
To examine the role of Otogl in barrier function, we employed a splice morpholino to deplete Otogl. We have previously shown the effectiveness of this morpholino at disrupting Otogl mRNA splicing (7), and here using Western blotting, we show its ability to reduce Otogl at the protein level (Fig. 3A). The blot shows clear loss of Otogl signal (both OG and OP) in lysate from Otogl morphants compared with controls when probed with both the Otogl antibody and PNA. PNA binding to the CG material was not altered and demonstrated equal loading between lanes.
Fig. 3.
Otogl forms a host-protective barrier on the epidermal surface. (A) Western blot of lysate from MOC- and Otogl MO-injected tadpoles, coprobing with the anti-Otogl antibody (green) and PNA (red), shows loss of Otogl protein upon knockdown. Glycosylated (OG) and precursor (OP) forms of Otogl are highlighted, while the similar intensity of signal for the cement gland mucin (CG) in the two lanes indicates equivalent loading of lysate. (B) Representative examples of sections from snap-frozen MOC- and Otogl MO-injected tadpoles stained with anti-Otogl antibody and DAPI. (Scale bar: 25 μm.) (C) Representative examples of sections from snap-frozen control MOC- and Otogl MO-injected tadpoles stained with PNA and DAPI. (Scale bar: 40 μm.) (D) Representative Cryo-TEM images of sections of snap-frozen MOC- and Otogl MO-injected tadpoles. Double-headed arrows show size of surface barrier. (Scale bar: 2 μm.) (E) Representative images of sections of MOC- and Otogl MO-injected tadpoles following exposure to GFP-expressing DH5α E. coli bacteria. White lines on images to the Left represent the apical surface membrane from brightfield images (Right). (Scale bar: 25 μm.)
To preserve and visualize the secreted barrier, we performed immunohistochemistry and lectin histochemistry on snap-frozen and cryo-sectioned tadpoles. Using the Otogl antibody and PNA, a layer was visualized on the skin surface of morpholino control (MOC)-injected tadpoles, which was depleted upon injection of the Otogl morpholino (Otogl MO) (Fig. 3 B and C). In subsequent experiments, PNA was used as a more sensitive probe for visualization of Otogl in the surface barrier.
To investigate the role of Otogl in forming a surface barrier, we used cryo-TEM on snap-frozen tadpoles injected with MOC or Otogl MO (Fig. 3D). MOC-injected tadpoles showed the presence of a continuous surface layer of ∼6 μm, whereas Otogl morphants had a thinner, less continuous surface layer (900 nm). We also conducted environmental scanning EM (ESEM) on live MOC and Otogl MO-injected tadpoles (Fig. S4). A smooth surface layer was evident in controls, with globular protrusions visible in a punctate pattern (boxes in Fig. S4). In Otogl morphants, the globular protrusions were much less visible, and there was no consistently smooth layer over the surface. Instead, the features and texture of the outer membrane could be observed. These data support the hypothesis that Otogl contributes to the structural integrity of a mucus barrier on the tadpole skin surface and that, when it is depleted, the tadpole epidermal cells are more exposed to the environment.
To test barrier function in Otogl morphants compared with controls, tadpoles were mixed with an overnight culture of Escherichia coli strain DH5α(pCoc2) that constitutively expresses GFP (19) and snap-frozen. Sections were taken and imaged as shown for representative examples in Fig. 3E. In MOC-treated tadpoles, GFP-expressing DH5α(pCoC2) bacteria were apparently “trapped” in the Otogl (marked by PNA) barrier, and few interacted directly with the underlying membrane. However, with Otogl morphants, the absence of an effective barrier allowed interaction of bacteria with the surface, and some bacteria were observed below the apical membrane (white arrows, Fig. 3E), although this was not specifically quantified.
Otogl Depletion Increases Sensitivity to Infection with an Opportunistic Pathogen, Aeromonas hydrophila.
Based on these findings, we reasoned that Otogl morphants would be compromised in terms of barrier protection and therefore hypothesized that these morphants would be more sensitive to infection than control animals. To test the functional impact of depleting Otogl (and thus the barrier), we infected control and Otogl morphant tadpoles with Aeromonas hydrophila, a bacterial species commonly found in freshwater environments that infects immunocompromised amphibians (20).
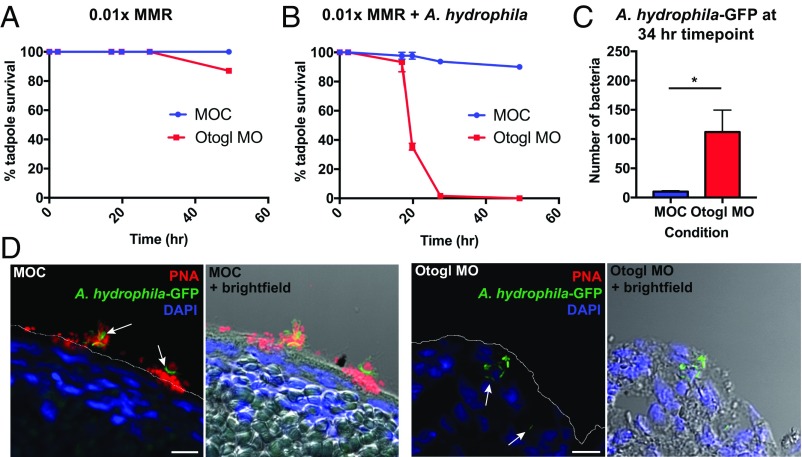
Controls and Otogl morphants at 2 d postfertilization (2 d.p.f.) were infected with A. hydrophila at a dose of 1.5 × 108 cfu/mL. This dose was experimentally confirmed in a preliminary experiment to identify the highest dose of A. hydrophila in which wild-type tadpoles could survive and thrive (e.g., swim and respond to touch stimuli). Before conducting the time-course infection experiments, MOC- and Otogl MO-injected animals (at 2 d.p.f.) were assessed for survival without A. hydrophila infection (Fig. 4A). Tadpoles were raised for a further 48 h, and survival levels were recorded at individual time points. In this experiment, there was no difference in survival between control and Otogl MO animals in the first 24 h, and even after 48 h, 80% of Otogl morphants were alive compared with all of the control animals.
Fig. 4.
Otogl morphants are sensitive to infection with A. hydrophila. (A) Survival time course of MOC-injected and Otogl MO-injected tadpoles in 0.01× Marc’s Modified Ringer’s (MMR). (B) Survival time course of MOC-injected and Otogl MO-injected tadpoles in 0.01× MMR containing 1.5 × 108 cfu/mL of A. hydrophila (at time point 0 h). Individual points represent mean survival levels from three independent experiments, and error bars represent the SEM. (C) Bar chart comparing the frequency of GFP-expressing A. hydrophila bacteria located within the tadpole in MOC- and Otogl MO-injected tadpoles fixed and sectioned at the 34-h time point. Bars represent mean number of bacteria found within MOC (n = 3 tadpoles)- and Otogl MO (n = 5 tadpoles)-injected tadpoles. Error bars represent SEM and P = 0.0179 (one-tailed Mann–Whitney U test). (D) Representative images of sections of MOC- and Otogl MO-injected tadpoles following exposure to GFP-expressing A. hydrophila bacteria at the 34-h time point. The white line on the Left image represents the apical surface membrane from brightfield images (Right). (Scale bar: 10 μm.)
The A. hydrophila infection experiments were repeated in three independent experiments, and the mean results recorded as shown in Fig. 4B. There was a slight decrease in survival levels of Otogl morphants compared with controls after 17 h, but this became much more pronounced by 20 h, with all Otogl morphant tadpoles dead after 27 h, compared with over 90% of control animals still alive (Student t test, P < 0.0001). By the end of the 48-h time course, ∼90% of controls still were alive, swimming and responsive to touch. To test if the reduced survival level of Otogl morphants was due to the impact of live A. hydrophila bacteria as opposed to a host response to bacterial products, we repeated the survival time course with heat-killed A. hydrophila (Fig. S5A). The Otogl morphants did not show the decline in survival with heat-killed bacteria that was observed with live bacteria, indicating that reduced survival was due to the actions of viable bacteria on the tadpole.
To demonstrate that the reduced survival levels of Otogl morphants was due to increased bacterial entry into the tadpoles, we generated GFP-expressing A. hydrophila and repeated the infection time course, fixing tadpoles at early and late time points and counting numbers of bacteria inside the tadpole (Fig. 4 C and D and Fig. S5 B–D). In terms of the tadpole survival time course, the same trend as for untransformed A. hydrophila was observed for the GFP-expressing variant, with Otogl morphants showing reduced survival compared with controls starting after ∼18 h postinfection (Fig. S5B). Fixation and sectioning of tadpoles at both an early (18 h 30 min) and late (34 h) time point, we compared the numbers of A. hydrophila-GFP bacteria inside the tadpoles by staining with a GFP antibody. There was no significant difference between the numbers of bacteria identified inside the Otogl morphant tadpoles compared with controls at 18 h 30 min, with very few bacteria recorded in each case (Fig. S5C). However, by 34 h, significantly more (one-tailed Mann–Whitney U test, P < 0.05) bacteria were counted inside the tadpoles of Otogl morphants compared with controls (Fig. 4C). Representative images at the 34-h time point show GFP-expressing bacteria located within the tadpole in Otogl morphants, while controls show bacteria outside, often in contact with Otogl on the skin surface (white arrows, Fig. 4D). Furthermore, the increased load of GFP-expressing A. hydrophila inside Otogl morphants was often accompanied by signs of necrosis such as changes in cell shape, size, and hyperpigmentation (Fig. S5). The latter is most likely due to recruitment of melanocytes, which have been shown to be important in innate immune defense of the tadpole after infection (21). Overall, the results show that the physical barrier generated by Otogl is critical for protection against infection.
Discussion
This study demonstrates that genetic knockdown of Otogl leads to depletion in the X. tropicalis tadpole skin-surface physical barrier that results in an increased susceptibility to infection. This reflects what has been shown for mammalian gel-forming mucins. In the mammalian respiratory tract, knocking out the mouse respiratory gel-forming mucin Muc5b (22) results in a major defect in mucociliary clearance, resulting in obstruction of the airways and increased susceptibility to infection by multiple bacterial species. In the intestine, mice deficient in Muc2, the predominant intestinal mucin (23), have a depleted mucus barrier that leads to enhanced susceptibility to infection with Citrobacter rodentium (24) and Salmonella enterica (25). Moreover, Muc2-deficient mice have impaired resistance to enteric parasitic infection (26). Like the frog, other aquatic organisms such as fish produce mucus at their skin surface to protect against environmental pathogens. Indeed, physical removal of mucus from the surface of channel catfish leads to increased susceptibility to opportunistic infection by A. hydrophila (27). These studies all show how important physical mucus barriers are for host protection, and the experimentally tractable tadpole model employed here provides a platform to define the key molecular aspects of the interplay between the host and invading microbes.
We have shown that Otogl bears a strong similarity to gel-forming mucins in sequence, domain organization, O-glycosylation, and structural properties, as well as in the capacity to form a protective barrier. As you would predict for a member of the mucin family, which is remarkably diverse both within and between species (28, 29), there are some interesting and novel differences between Otogl and the major mammalian gel-forming mucins.
Specifically, the central mucin domain is typically encoded by one single long exon in mammalian mucins, whereas it is multiexonic in Otogl. There is a precedence for this; the chicken mucin, Muc13, has multiple exons encoding its central domain (30). Otogl does not contain typical CysD regions often found in the mucin domain of gel-forming mucins (1). Instead of CysD domains, Otogl has multiple Cys-rich domains that may perform a similar role in mediating intermolecular interactions and altering the pore size of mucus (14).
As a result of the data presented here, it is clear that Otogl is actually a gel-forming mucin rather than a member of the otogelin family and as such is misnamed. Although otogelins are evolutionarily related to mucins containing VWD domains (28), there are key differences. Specifically, otogelins lack the extensive central domain that is rich in proline, threonine, and serine residues and that is predominantly O-glycosylated. In contrast, Otogelins are mainly N-glycosylated (12). Finally, and perhaps most pertinently, Otogl is not expressed in the inner ear as are other otogelin molecules and instead forms a physical barrier to infection like all known gel-forming mucins. As such we propose that its name should change to reflect its function. We advocate the name Xenopus Skin Mucin, or more succinctly, MucXS.
Analysis of the X. tropicalis genome has shown that at least 26 gel-forming mucins are expressed at varying levels at different stages of development (27, 28). Multiple variants of Muc2 and Muc5 and single variants of Muc6 and Muc19 have been identified. As with MucXS, these mucins share central PTS-rich regions interrupted by Cys-rich sequences, N- and C-terminal VWD domains, and a C-terminal CK domain. MucXS was not identified in these studies likely due to the lack of sequence for the central region that we have described here.
In conclusion, the data presented in the current study clearly demonstrate that X. tropicalis tadpoles form a physical host-protective barrier to infection at their skin surface that is underpinned by a secreted gel-forming mucin, MucXS. Our analysis shows that MucXS has a critical role in mediating host–pathogen interactions. Establishing the presence of this mucin-based barrier advances the potential of this model system for studying the fundamental biology of the mucus barrier and host–microbe interactions at mucosal epithelia. Importantly, we have previously identified a number of other proteins secreted from the tadpole skin that are conserved in mammalian mucus, such as lectins and the mucin-related protein IgG Fcγ-binding protein (7). Now that we have characterized MucXS, it remains to be investigated how all these components interact together to build the barrier and promote host defense. Because this simple tadpole system is external and easily accessible to manipulation, as well as live imaging, it is a unique in vivo model in comparative biology to study host–pathogen interactions at mucosal surfaces.
Materials and Methods
Fluorescence in Situ Hybridization.
Clones TNeu027a13 and ThdA045k18 were used to generate digoxygenin-labeled RNA probes; see SI Materials and Methods for detailed experimental protocols. Fluorescence in situ hybridization was performed on tadpoles as described previously (31).
Sequencing of Otogl.
cDNA was generated using SuperScript IV Reverse Transcriptase (ThermoFisher). Long-template PCR was performed on the cDNA using Phusion High Fidelity DNA Polymerase (NEB); see SI Materials and Methods for primer sequences. PCR products were gel-extracted, cloned into the pCR-XL-TOPO vector, and sequenced using the Pacific Biosciences SMRT sequencing platform at the Earlham Institute, Norwich, UK. See SI Materials and Methods for detailed experimental protocols.
Morpholino Design and Injection.
All morpholinos were purchased from Gene Tools. The otogl splice morpholino sequence is 5′-TAGAGTCATACATACCTCCATCATC-3′. The control morpholino sequence is 5′- CCTCTTACCTCAGTTACAATTTATA-3′. Morpholinos were injected with a 15-ng dose at the one-cell stage.
Purification and Characterization of Otogl.
Secretions were collected from 5,000 wild-type tadpoles, and lysates were collected in lysis buffer containing protease inhibitors. Otogl was purified from secretions by 4 M GdmCl/CsCl density-gradient centrifugation, and its identity was confirmed by tandem mass spectrometry. The size distribution of purified Otogl was analyzed by rate-zonal centrifugation (2), its molecular weight was analyzed by SEC-MALLS (13), and its macromolecular form was visualized by TEM. Glycosylation of Otogl was analyzed by lectin- (PNA) and immunoblotting (anti-Otogl) after agarose electrophoresis; samples were digested with PNGase F, O-glycosidase, and/or sialidase. The cellular localization of Otogl was determined by immuno- and lectin fluorescence microscopy on whole tadpoles using anti-Otogl and PNA. Membrane GFP was used to mark cell boundaries (7). See SI Materials and Methods for detailed experimental protocols.
Visualization of the Skin-Surface Mucus Barrier.
Ultrathin sections of liquid-ethane–frozen tadpoles were visualized by cryo-TEM; see SI Materials and Methods for the detailed experimental protocol. Surface ESEM imaging of live tadpoles was performed on an FEI Model Quanta 200 ESEM. Immunofluorescence microscopy of the surface barrier was performed on liquid N2 snap-frozen tadpoles (32, 33); see SI Materials and Methods for the detailed experimental protocols.
Infection of Tadpoles with E. coli DH5α(pCOC2)-GFP and A. hydrophila.
Control and Otogl morphant tadpoles were incubated with E. coli DH5α(pCOC2)-GFP (19), A. hydrophila (ATCC7966), or GFP-expressing A. hydrophila. See SI Materials and Methods for the detailed experimental protocols.
Supplementary Material
Acknowledgments
We thank Dr. Jonathan Shaw (University of Sheffield) for the kind gift of A. hydrophila ATCC7966; Marie Goldrick for assistance with the bacterial experiments; Patrick Hill for assistance with ESEM; Professor Enrique Amaya and Professor Nancy Papalopulu for use of Xenopus and laboratory facilities; and the Biomolecular Analysis, Bio-MS, and EM core facilities (University of Manchester). This research was supported by funding from the Biotechnology and Biological Sciences Research Council (Grant BB/M021688/1). The Wellcome Trust Centre for Cell-Matrix Research, University of Manchester, is supported by core funding from the Wellcome Trust (Grant 203128/Z/16/Z).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. MG724746).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713539115/-/DCSupplemental.
References
- 1.Thornton DJ, Rousseau K, McGuckin MA. Structure and function of the polymeric mucins in airways mucus. Annu Rev Physiol. 2008;70:459–486. doi: 10.1146/annurev.physiol.70.113006.100702. [DOI] [PubMed] [Google Scholar]
- 2.Thornton DJ, et al. Mucus glycoproteins from ‘normal’ human tracheobronchial secretion. Biochem J. 1990;265:179–186. doi: 10.1042/bj2650179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radicioni G, et al. The innate immune properties of airway mucosal surfaces are regulated by dynamic interactions between mucins and interacting proteins: The mucin interactome. Mucosal Immunol. 2016;9:1442–1454. doi: 10.1038/mi.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson AG, et al. Cystic fibrosis airway secretions exhibit mucin hyperconcentration and increased osmotic pressure. J Clin Invest. 2014;124:3047–3060. doi: 10.1172/JCI73469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johansson MEV, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubaissi E, Papalopulu N. Embryonic frog epidermis: A model for the study of cell-cell interactions in the development of mucociliary disease. Dis Model Mech. 2011;4:179–192. doi: 10.1242/dmm.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubaissi E, et al. A secretory cell type develops alongside multiciliated cells, ionocytes and goblet cells, and provides a protective, anti-infective function in the frog embryonic mucociliary epidermis. Development. 2014;141:1514–1525. doi: 10.1242/dev.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walentek P, Quigley IK. What we can learn from a tadpole about ciliopathies and airway diseases: Using systems biology in Xenopus to study cilia and mucociliary epithelia. Genesis. 2017;55:e23001. doi: 10.1002/dvg.23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagata S, Nakanishi M, Nanba R, Fujita N. Developmental expression of XEEL, a novel molecule of the Xenopus oocyte cortical granule lectin family. Dev Genes Evol. 2003;213:368–370. doi: 10.1007/s00427-003-0341-9. [DOI] [PubMed] [Google Scholar]
- 10.Walentek P, et al. A novel serotonin-secreting cell type regulates ciliary motility in the mucociliary epidermis of Xenopus tadpoles. Development. 2014;141:1526–1533. doi: 10.1242/dev.102343. [DOI] [PubMed] [Google Scholar]
- 11.Hayes JM, et al. Identification of novel ciliogenesis factors using a new in vivo model for mucociliary epithelial development. Dev Biol. 2007;312:115–130. doi: 10.1016/j.ydbio.2007.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen-Salmon M, El-Amraoui A, Leibovici M, Petit C. Otogelin: A glycoprotein specific to the acellular membranes of the inner ear. Proc Natl Acad Sci USA. 1997;94:14450–14455. doi: 10.1073/pnas.94.26.14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridley C, et al. Assembly of the respiratory mucin MUC5B: A new model for a gel-forming mucin. J Biol Chem. 2014;289:16409–16420. doi: 10.1074/jbc.M114.566679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambort D, et al. Function of the CysD domain of the gel-forming MUC2 mucin. Biochem J. 2011;436:61–70. doi: 10.1042/BJ20102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridley C, et al. Biosynthesis of the polymeric gel-forming mucin MUC5B. Am J Physiol Lung Cell Mol Physiol. 2016;310:L993–L1002. doi: 10.1152/ajplung.00046.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao R, Wang TT, DeMaria G, Sheehan JK, Kesimer M. Mapping the protein domain structures of the respiratory mucins: A mucin proteome coverage study. J Proteome Res. 2012;11:4013–4023. doi: 10.1021/pr300058z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Vilar J, Hill RL. The structure and assembly of secreted mucins. J Biol Chem. 1999;274:31751–31754. doi: 10.1074/jbc.274.45.31751. [DOI] [PubMed] [Google Scholar]
- 18.Kesimer M, Makhov AM, Griffith JD, Verdugo P, Sheehan JK. Unpacking a gel-forming mucin: A view of MUC5B organization after granular release. Am J Physiol Lung Cell Mol Physiol. 2010;298:L15–L22. doi: 10.1152/ajplung.00194.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corcoran CP, Cameron ADS, Dorman CJ. H-NS silences gfp, the green fluorescent protein gene: gfpTCD is a genetically remastered gfp gene with reduced susceptibility to H-NS-mediated transcription silencing and with enhanced translation. J Bacteriol. 2010;192:4790–4793. doi: 10.1128/JB.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mauel MJ, Miller DL, Frazier KS, Hines ME., II Bacterial pathogens isolated from cultured bullfrogs (Rana castesbeiana) J Vet Diagn Invest. 2002;14:431–433. doi: 10.1177/104063870201400515. [DOI] [PubMed] [Google Scholar]
- 21.Paré J-F, Martyniuk CJ, Levin M. Bioelectric regulation of innate immune system function in regenerating and intact Xenopus laevis. Npj Regen Med. 2017;2:1–14. doi: 10.1038/s41536-017-0019-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy MG, et al. Muc5b is required for airway defence. Nature. 2014;505:412–416. doi: 10.1038/nature12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergstrom KSB, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarepour M, et al. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect Immun. 2013;81:3672–3683. doi: 10.1128/IAI.00854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasnain SZ, et al. Mucin gene deficiency in mice impairs host resistance to an enteric parasitic infection. Gastroenterology. 2010;138:1763–1771. doi: 10.1053/j.gastro.2010.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, et al. Evasion of mucosal defenses during Aeromonas hydrophila infection of channel catfish (Ictalurus punctatus) skin. Dev Comp Immunol. 2013;39:447–455. doi: 10.1016/j.dci.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Lang T, Hansson GC, Samuelsson T. Gel-forming mucins appeared early in metazoan evolution. Proc Natl Acad Sci USA. 2007;104:16209–16214. doi: 10.1073/pnas.0705984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang T, et al. Searching the evolutionary origin of epithelial mucus protein components-mucins and FCGBP. Mol Biol Evol. 2016;33:1921–1936. doi: 10.1093/molbev/msw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang T, Hansson GC, Samuelsson T. An inventory of mucin genes in the chicken genome shows that the mucin domain of Muc13 is encoded by multiple exons and that ovomucin is part of a locus of related gel-forming mucins. BMC Genomics. 2006;7:197. doi: 10.1186/1471-2164-7-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lea R, Bonev B, Dubaissi E, Vize PD, Papalopulu N. Multicolor fluorescent in situ mRNA hybridization (FISH) on whole mounts and sections. Methods Mol Biol. 2012;917:431–444. doi: 10.1007/978-1-61779-992-1_24. [DOI] [PubMed] [Google Scholar]
- 32.Dubaissi E, Panagiotaki N, Papalopulu N, Vize PD. In: Antibody Development and Use in Chromogenic and Fluorescent Immunostaining. Xenopus Protocols, Methods in Molecular Biology. Hoppler S, Vize PD, editors. Humana Press; Totowa, NJ: 2012. pp. 411–429. [DOI] [PubMed] [Google Scholar]
- 33.Cohen M, Varki NM, Jankowski MD, Gagneux P. Using unfixed, frozen tissues to study natural mucin distribution. J Vis Exp. 2012:e3928. doi: 10.3791/3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miles AA, Misra SS, Irwin JO. The estimation of the bactericidal power of the blood. J Hyg (Lond) 1938;38:732–749. doi: 10.1017/s002217240001158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortinea N, et al. Optimization of green fluorescent protein expression vectors for in vitro and in vivo detection of Listeria monocytogenes. Res Microbiol. 2000;151:353–360. doi: 10.1016/s0923-2508(00)00158-3. [DOI] [PubMed] [Google Scholar]
- 36.Cohen SN, Chang AC, Hsu L. Nonchromosomal antibiotic resistance in bacteria: Genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci USA. 1972;69:2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.