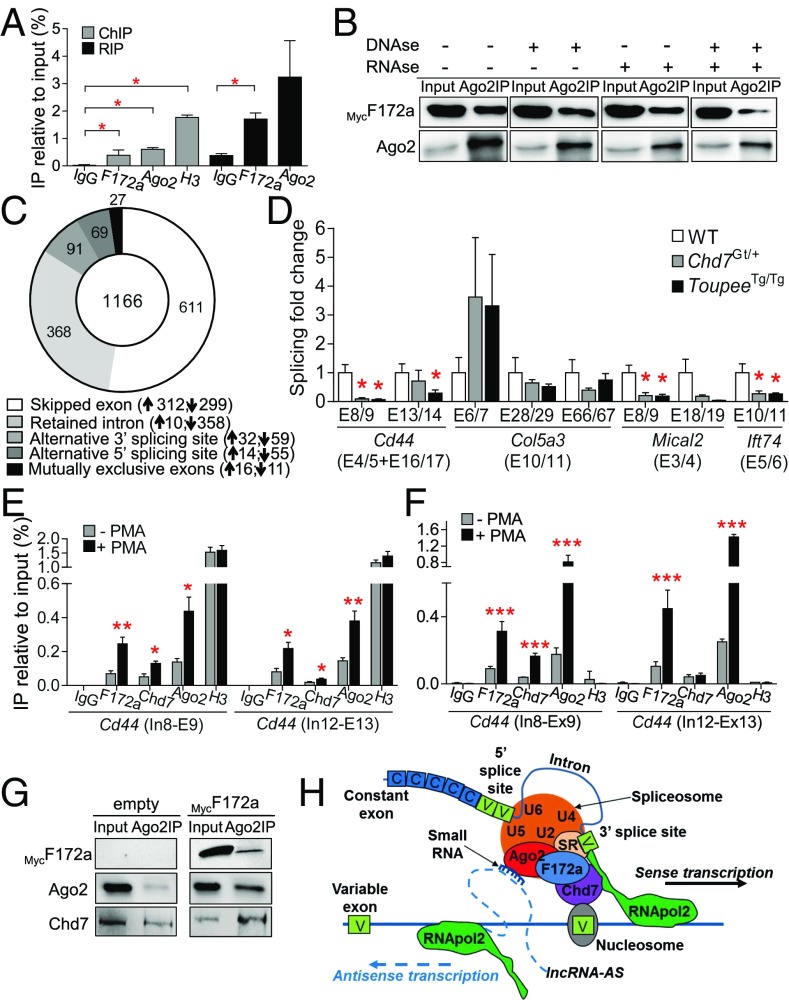
Fig. 4.
Both Fam172a and Chd7 play a role in alternative splicing. (A) Untargeted ChIP and RIP assays in Neuro2A cells transfected with a Fam172a-expressing plasmid (n = 3). (B) Co-IP assays using RNase- and/or DNase-treated whole-cell extracts of Neuro2A cells transfected with a MycFam172a expression vector. Inputs correspond to 10% of protein extracts used for IP (n = 3 per condition). Impact of each treatment on the integrity of proteins, DNA, and RNA is shown in SI Appendix, Fig. S8G. (C) Donut chart showing the distribution of the 1,166 differentially modulated alternative splicing events (P < 0.01; variation in inclusion level ≥0.1) in ToupeeTg/Tg E10.5 NCCs. Upward- and downward-pointing arrows indicate splicing events that are over- and under-represented in ToupeeTg/Tg E10.5 NCCs, respectively. (D) RT-qPCR analysis of splicing events for Cd44, Col5a3, Mical2, and Ift74 in G4-RFP (WT), ToupeeTg/Tg;G4-RFP (ToupeeTg/Tg), and Chd7Gt/+ heads of E12.5 embryos (n = 5 per genotype). Expression levels of variable regions are normalized with levels of corresponding constant regions (indicated between parentheses). (E and F) ChIP (E) and RNA-ChIP (F) assays of the PMA-inducible Cd44 gene in Neuro2a cells. (G) Co-IP assays in Neuro2A cells transfected with empty or MycFam172a-expressing vector. Inputs correspond to 10% of protein extracts used for IP (n = 3 per condition). (H) Potential mode of action of Fam172a and Chd7 in Ago2-mediated alternative splicing (adapted from ref. 21). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (Student’s t test).