Figure 1.
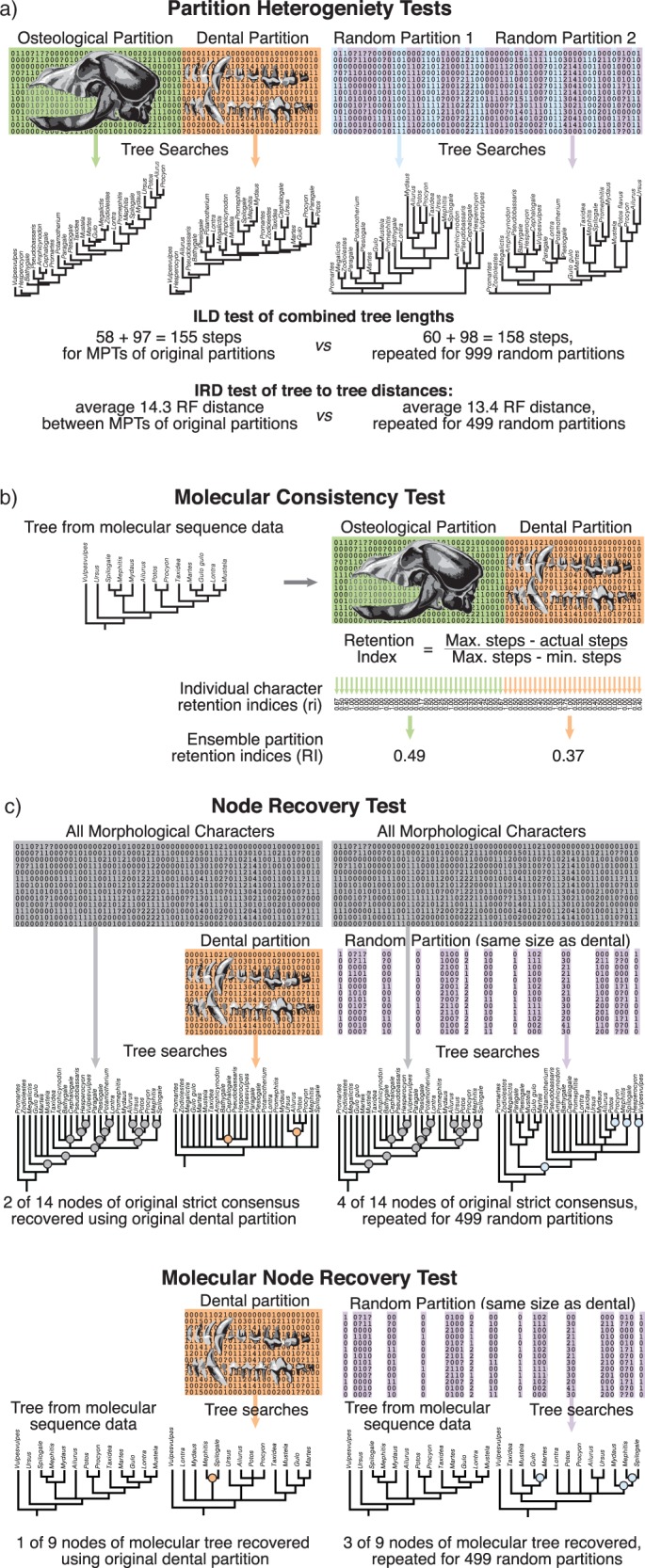
Analyses applied to morphological data. a) Partition heterogeneity tested by comparing combined tree length (ILD) or nearest neighbor tree-to-tree distances of trees (IRD) resulting from searches of each morphological character partition (osteological and dental) and random character partitions of the same size (Farris et al. 1995a, 1995b; Mounce et al. 2016). b) Molecular consistency tested by applying morphological data to a molecular tree and calculating retention indices of characters and partitions. c) Node recovery tested by comparing strict consensus trees resulting from searches using only dental characters and strict consensus trees resulting from searches using random subsets of characters in the same proportion (i.e., loss of signal with either systematic or random character removal). Signal recovery is assessed in terms of nodes shared with either the strict consensus tree using all characters or the molecular tree. Morphological data set example from Finarelli (2008) with Flynn et al. (2005) molecular tree.
