Table 2.
Selected 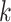 -mer block algorithm parameter
settings and resulting data set characteristics obtained for each genome data
set
-mer block algorithm parameter
settings and resulting data set characteristics obtained for each genome data
set
| Clade |
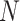
|
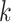 (index
(index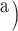
|
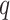
|
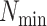
|
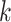 -mer blocks
-mer blocks |
Alignment length |
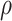
|
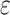
|
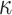
|
|---|---|---|---|---|---|---|---|---|---|
| Land plant mitochondrial | 93 | 24 (0) | 2 | 10 | 2279 | 145,856 | 0.25 |
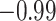
|
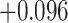
|
| Angiosperm plastid | 663 | 28 (5) | 2 | 100 | 1711 | 116,348 | 0.51 |
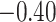
|
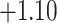
|
| Caryophyllales | 67 | 28 (0) | 2 | 15 | 684 | 46,512 | 0.58 |
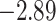
|
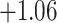
|
| Oryza | 11 | 32 (0) | 0 | 11 | 35,646 | 2,566,512 | 1.00 |
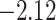
|
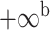
|
| Anopheles | 17 | 32 (0) | 2 | 12 | 38,648 | 2,782,656 | 0.89 |
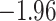
|
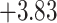
|
| Fabaceae | 8 | 32 (1) | 2 | 4 | 33,609 | 2,419,848 | 0.82 |
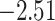
|
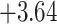
|
| Eudicots | 24 | 28 (0) | 2 | 5 | 31,897 | 2,168,996 | 0.36 |
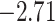
|
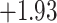
|
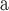 Index indicates the numeral in the supermatrix file name
corresponding to this run and these parameters (see Supplementary Materials
available on Dryad);
Index indicates the numeral in the supermatrix file name
corresponding to this run and these parameters (see Supplementary Materials
available on Dryad); 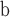 Undefined, because
Undefined, because
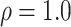 .
.
