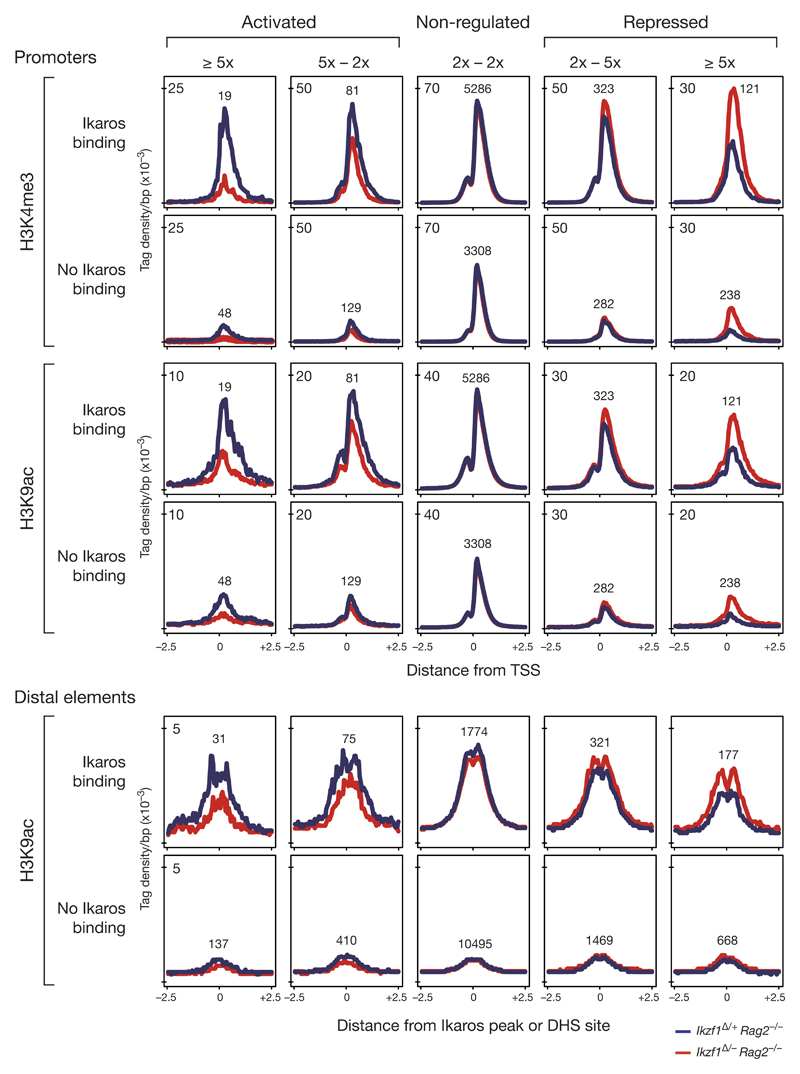
Figure 7. Chromatin changes at promoters and distal elements of Ikaros-regulated genes.
Ikaros-dependent chromatin changes were identified by ChIP-seq mapping of the active histone modifications H3K4me3 and H3K9ac in short-term cultured Cd79a-Cre Ikzf1fl/– Rag2–/– (red) and Cd79a-Cre Ikzf1fl/+ Rag2–/– (blue) pro-B cells. Genes were classified according to their degree of Ikaros-dependent regulation (in the two Rag2–/– pro-B cell types) and the presence or absence of Ikaros-binding sites at their regulatory elements. The average density of the histone marks is shown for a region extending from -2.5 kb to +2.5 kb relative to the TSS at promoters or center of the Ikaros peak or DHS site (no Ikaros binding) at distal elements. Only promoters and distal elements of genes with an expression level of > 2 RPKM in control pro-B cells (activated, not regulated) or in Ikaros-deficient pro-B cells (repressed) were analyzed. The number of peaks analyzed is shown for each category. Wilcoxon pair-matched comparison of the peak density at the regulatory elements in Ikzf1Δ/– Rag2–/– and Ikzf1Δ/+ Rag2–/– pro-B cells revealed p-values of < 10-3 for all chromatin changes observed at promoters and distal elements of activated (> 2x) and repressed (> 2x) genes.