ABSTRACT
The root cap protects the root from environmental stress and senses gravity. Cells of the last layer of the root cap are shed in a developmentally programmed process. We previously showed that the transcription factor NIN-LIKE PROTEIN7 (NLP7) regulates root cap cell release likely through regulation of CELLULASE5 (CEL5). Here we provide a supplement to that work. We hypothesized that the nlp7 mutant has defects in additional root cap functions. We find that neither gravity sensing nor expression of a root cap cell identity marker is altered in nlp7 but that expression of another cellulase, CEL3, is upregulated. We conclude that NLP7 control of root cap cell release is largely independent of gravity sensing and root cap cell identity.
KEYWORDS: Columella, cell release, NLP7, NIN transcription factors, root cap
Roots are key for plant health and fitness, as they uptake water and nutrients, anchor plants in soil, and protect plants against soil borne pathogens. The root cap, a specialized tissue at the tip of the root, is critical for a root's ability to navigate the belowground environment. The root cap protects the root meristem,1 particularly from biotic stress,2 senses gravity and mechanical obstacles,3,4 and is important for hydrotropism.5,6 The Arabidopsis root cap is composed of primarily two distinct cell types, the lateral root cap and the columella root cap (COL). In Arabidopsis, the central part of the root cap is composed of rectangular COL cells. As COL cells mature, new cells are produced from COL initials while cells in the last layer of the root cap are released from the root, in a process that keeps the COL a stable size.7
In most plant families other than the Brassicaceae, the outermost cells of the root cap are known as border cells (BCs), and are released as single cells.1 However, in several species of Brassicaceae, border cells are attached to one another even after they are released from the root cap and are called Border Like Cells (BLCs). BCs and BLCs secrete mucilaginous substances with antimicrobial compounds and secondary metabolites that defend the root against soil-borne pathogens. The mucilage also plays an important role in biotic stress tolerance.8-11 The mucilage produced by BCs and BLCs may reduce mechanical resistance as the root grows through soil.1 The release of BLC depends on the action of enzymes that modify components of the cell wall including cellulases and pectin methylesterases.12-15 Mutants deficient in cell- wall- homogalacturonan (HG) shows defects in the release of BLCs, with single cells released from the root cap instead of an intact layer.12,14,15 BLC release is also influenced by degradation of cellulose by cellulases, as a mutant defective in CELLULASE5 (CEL5) shows a sticky root cap.13,15
We have recently shown that the transcription factor NIN-LIKE PROTEIN 7 (NLP7), previously characterized as a regulator of nitrate signaling,16-18 modulates BLC release in Arabidopsis.15 Mutations in NLP7 lead to the release of BLCs as single cells and to the upregulation of several cell wall modifying enzymes including CELLULASE5.15 The nlp7 BLC release phenotype is dependent upon a functional CEL5, because the nlp7-1cel5 double mutant releases BLCs as an intact layer.
We hypothesized that due to the defects in root cap cell release, the nlp7-1 mutant would have other defects in the root cap, such as altered gravity sensing or COL cell identity. Here we show that gravity sensing is not altered in the nlp7-1 mutant. In addition, expression of a root cap cell identity marker in the nlp7-1 background is similar to wild type. Finally, we show that another cellulase, CEL3, is upregulated in roots of nlp7-1, but that mutations in CEL3 do not lead to altered root cap layers, possibly due to functional redundancy.
To examine whether columella root cap identity was altered, we crossed the previously characterized PET111:GFP enhancer trap line19 with nlp7-1. PET111:GFP is expressed in columella root cap cells19 and is frequently used as a marker for this cell type.20-22 Examination of 7 day old roots of nlp7-1 PET111:GFP revealed a similar expression pattern as that of wild type (WT) PET111:GFP (Fig. 1A, B). This suggests that although there is a reduction in the COL root cap layers in nlp7-1,15 the COL cellular identity is not affected.
Figure 1.
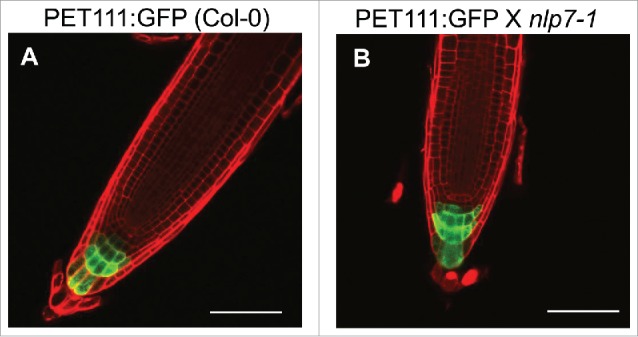
The columella root cap marker PET111 has similar expression in both Col-0 and nlp7-1. (A) PET111 expression in Col-0, (B) PET111 X nlp7-1. Scale bar = 100 µm.
We next asked whether root cap physiological functions were defective in nlp7-1. Apart from protecting the developing meristem, one major function of the root cap is sensing gravity. COL cells contain specialized starch grains called amyloplasts that play a crucial role in the gravity sensing mechanism of root cap. To examine a role for NLP7 in gravity sensing, we tested the effect of gravity on the nlp7-1 mutant. We grew WT and nlp7-1 seedlings vertically on agar plates for 7 days, and the plates were then turned 180 degrees. We measured the angle of root curvature of the WT and nlp7-1 24 hours after gravistimulation (Fig. 2A). As shown in Fig. 2A, no significant difference in the angle of root curvature between WT and nlp7-1 was observed after gravity treatment. Next, we examined the amyloplast content of the nlp7-1 mutant. Starch staining with 1% Lugol solution showed that COL cells of nlp7-1 contain amyloplasts with starch (Fig. 2B, C). These results indicate that NLP7 does not affect the gravity sensing mechanism of columella root cap cells.
Figure 2.
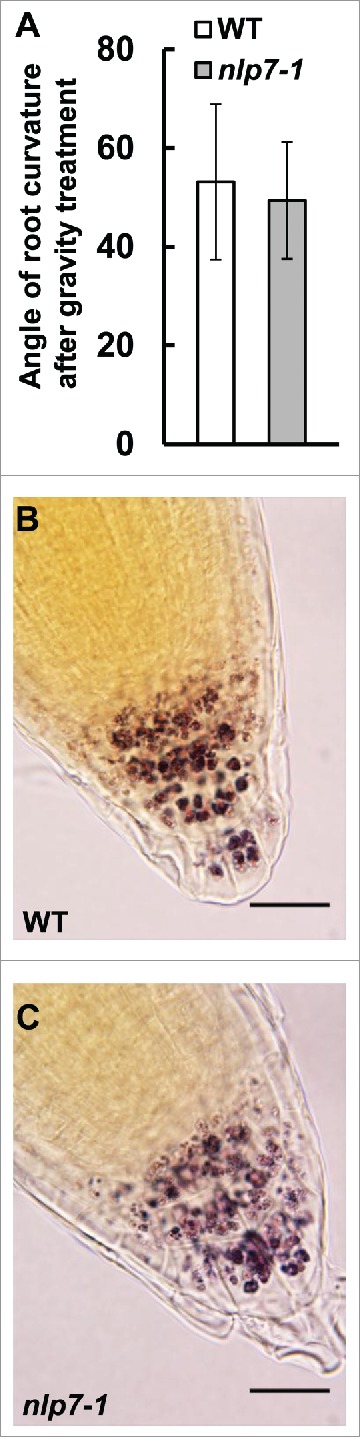
NLP7 does not regulate the gravity sensing mechanism of the root cap. (A) Measurement of root curvature angle 24 h after gravity treatment in WT and nlp7-1 mutant plants. No significant difference (P > 0.05) was observed between WT and nlp7-1. N = 18 or more roots. (B) and (C) Root caps of WT (B) and nlp7-1 (C) stained with 1% Lugol's solution to stain for starch. Images taken at 100X. Scale bar = 20 µm.
Together, these data suggest that the role of NLP7 in root cap cell release is independent of columella cell identity and gravity sensing. To identify additional cell wall enzymes with expression that may be modulated by NLP7, we focused on another gene in the CELLULASE family. We chose to focus on CEL3 (At1g71380) because it is highly expressed in the COL and lateral root caps.23 We studied the transcript levels of CEL3 in wild type and the nlp7-1 mutant and found that CEL3 expression is 2.5 fold higher in the nlp7-1 mutant (Fig. 3A), which is similar to the expression level of CEL5 in nlp7-1.15 Although a defect in CEL5 leads to additional root cap layers,15 examination of the cel3-1 mutant (Salk_057689) revealed no difference in the number of root cap layers between WT and cel3-1 (Fig. 3B), possibly due to redundancy in the CELLULASE gene family.
Figure 3.
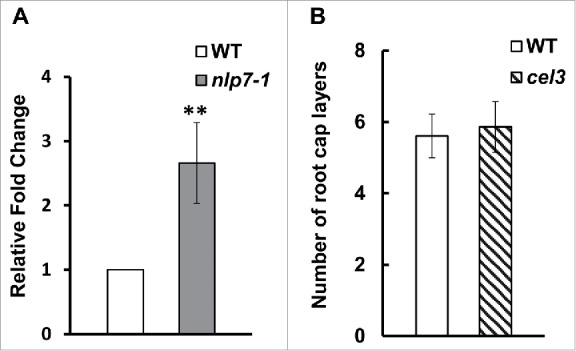
CEL3 is expression is upregulated in nlp7-1 mutant but mutations in CEL3 do not lead to a sticky root cap layer phenotype. (A) Quantitative PCR showing upregulation of CEL3 in nlp7-1 mutant. Error bars show standard deviation between three independent biological replicates. P < 0.05, (B) Number of root cap layers from columella initials to the last layer of border like cells. No significant difference was observed in the number of root cap layers between WT and the cel3-1 mutant. N = 20 roots per genotype.
We conclude that NLP7 has a distinct role in root cap cell release through the regulation of cell wall degrading enzymes and does not appear to function in columella root cap cell identity or gravity sensing.
Materials and methods
Plant materials and growth conditions
The PET111 enhancer trap line19 in wild type Columbia (Col-0) was crossed to the nlp7-1 mutant (Salk_026134). Homozygous nlp7-1 seedlings expressing the PET111 marker were sterilized in 50% bleach and stratified at 4°C for 48 h. Seeds were plated on 1X Murashige and Skoog (MS, Caisson Labs), pH 5.7 with 1% sucrose and 1% agar. Seedlings were grown vertically in a growth chamber at 22 – 23°C with 16 hour day length. Roots were observed at 7 days after germination with confocal microscopy as previously described.15
Homozygous seedlings of the cel3-1 mutant (Salk_057689) were sterilized and grown as above. The root cap was viewed with confocal microscopy at 7 days after germination as previously described. Root cap layers were counted as previously described.15
Lugol staining
Seedlings were grown on MS as above. At 5 days after germination, roots were stained for 1 minute in a 1% Lugol solution and placed immediately on a slide with Visikol (Visikol Inc, NJ, USA) as the mounting and clearing agent. Images were captured on an Olympus BX43 upright microscope under 100X magnification.
Gravistimulation assay
Seeds of WT (Col-0) and nlp7-1 were sterilized, stratified and plated on MS as above. Seedlings were grown vertically in a growth chamber. At 5 days after germination, plates were turned 180°. Plates were scanned 24 hours after turning and the angle between the primary root and the root hook formed from gravistimulation was measured using ImageJ.
Quantitative PCR
Seedlings were grown on MS as above. At 5 days after germination, RNA was extracted from whole roots as in.15 cDNA synthesis and qPCR were as in.15 Primers for CEL3 were: CEL3-qRT-F: 5′CAAGGTCAGCGATCTGGTCCTCTT 3′ and CEL3-qRT-R: 5′GGTGGTAAACGCCATTGGTAAATTG 3′ and the primer efficiency was evaluated with a cDNA dilution series. Relative expression was calculated relative to AT1G13320,24 using the delta-delta Ct method as in.15 Three biological replicates, each with three technical replicates, were performed.
Funding Statement
National Science Foundation (NSF) (1656392).
Disclosure of interest
The authors report no conflict of interest.
Acknowledgments
We thank Dr. Gloria Muday for providing cel3 mutant seeds. This work was supported by start-up funds from Purdue University and NSF grant 1656392.
References
- 1.Barlow P. The root cap: Cell dynamics, cell differentiation and cap function. J Plant Growth Regul. 2003;21:261–186. doi: 10.1007/s00344-002-0034-z. [DOI] [Google Scholar]
- 2.Hawes M, Allen C, Turgeon BG, Curlango-Rivera G, Minh Tran T, Huskey DA, Xiong Z. Root border cells and their role in plant defense. Annu Rev Phytopathol. 2016;54:143–161. doi: 10.1146/annurev-phyto-080615-100140. [DOI] [PubMed] [Google Scholar]
- 3.Blancaflor EB, Fasano JM, Gilroy S. Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol. 1998;116:213–22. doi: 10.1104/pp.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blancaflor EB, Fasano JM, Gilroy S. Laser ablation of root cap cells: implications for models of graviperception. Adv Space Res. 1999;24:731–8. doi: 10.1016/S0273-1177(99)00406-8. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi N, Yamazaki Y, Kobayashi A, Higashitani A, Takahashi H. Hydrotropism interacts with gravitropism by degrading amyloplasts in seedling roots of Arabidopsis and radish. Plant Physiol. 2003;132:805–10. doi: 10.1104/pp.018853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eapen D, Barroso ML, Ponce G, Campos ME, Cassab GI. Hydrotropism: root growth responses to water. Trends Plant Sci. 2005;10:44–50. doi: 10.1016/j.tplants.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Kumpf RP, Nowack MK. The root cap: a short story of life and death. J Exp Bot. 2015;66(19):5651–62. doi: 10.1093/jxb/erv295. [DOI] [PubMed] [Google Scholar]
- 8.Hawes MC, Gunawardena U, Miyasaka S, Zhao X. The role of root border cells in plant defense. Trends Plant Sci. 2000;5:128–33. doi: 10.1016/S1360-1385(00)01556-9. [DOI] [PubMed] [Google Scholar]
- 9.Vicre M, Santaella C, Blanchet S, Gateau A, Driouich A. Root border-like cells of Arabidopsis. Microscopical characterization and role in the interaction with rhizobacteria. Plant Physiol. 2005;138:998–1008. doi: 10.1104/pp.104.051813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Driouich A, Follet-Gueye ML, Vicre-Gibouin M, Hawes M. Root border cells and secretions as critical elements in plant host defense. Curr Opin Plant Biol. 2013;16:489–95. doi: 10.1016/j.pbi.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Watson BS, Bedair MF, Urbanczyk-Wochniak E, Huhman DV, Yang DS, Allen SN, Li W, Tang Y, Sumner LW. Integrated metabolomics and transcriptomics reveal enhanced specialized metabolism in Medicago truncatula root border cells. Plant Physiol. 2015;167:1699–716. doi: 10.1104/pp.114.253054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouton S, Leboeuf E, Mouille G, Leydecker MT, Talbotec J, Granier F, Lahaye M, Höfte H, Truong HN. QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell. 2002;14:2577–90. doi: 10.1105/tpc.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Campillo E, Abdel-Aziz A, Crawford D, Patterson SE. Root cap specific expression of an endo-beta-1,4-D-glucanase (cellulase): a new marker to study root development in Arabidopsis. Plant Mol Biol. 2004;56:309–23. doi: 10.1007/s11103-004-3380-3. [DOI] [PubMed] [Google Scholar]
- 14.Durand C, Vicre-Gibouin M, Follet-Gueye ML, Duponchel L, Moreau M, Lerouge P, Driouich A. The organization pattern of root border-like cells of Arabidopsis is dependent on cell wall homogalacturonan. Plant Physiol. 2009;150:1411–21. doi: 10.1104/pp.109.136382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karve R, Suarez-Roman F, Iyer-Pascuzzi AS. The transcription factor NIN-LIKE PROTEIN7 controls border-like cell release. Plant Physiol. 2016;171:2101–11. doi: 10.1104/pp.16.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, et al.. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009;57:426–35. doi: 10.1111/j.1365-313X.2008.03695.x. [DOI] [PubMed] [Google Scholar]
- 17.Konishi M, Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun. 2013;4:1617. doi: 10.1038/ncomms2621. [DOI] [PubMed] [Google Scholar]
- 18.Marchive C, Roudier F, Castaings L, Brehaut V, Blondet E, Colot V, Meyer C, Krapp A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun 2013; 4:1713. doi: 10.1038/ncomms2650. [DOI] [PubMed] [Google Scholar]
- 19.Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17:1908–25. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–6. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 21.Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–5. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- 22.Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN. Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell. 2011;21:770–82. doi: 10.1016/j.devcel.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis DR, Olex AL, Lundy SR, Turkett WH, Fetrow JS, Muday GK. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. Plant Cell. 2013;25:3329–46. doi: 10.1105/tpc.113.114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 2004;38:366–79. doi: 10.1111/j.1365-313X.2004.02051.x. [DOI] [PubMed] [Google Scholar]


