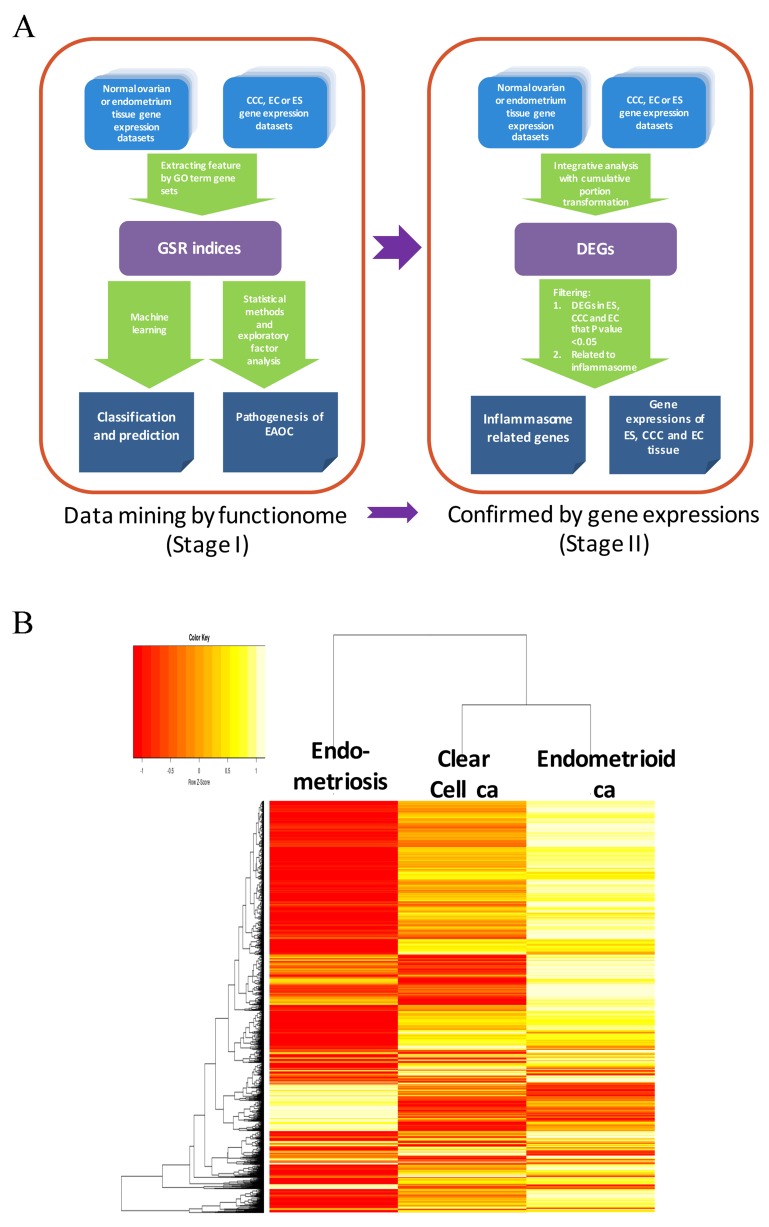
Figure 1. Work flow of the two-stage strategy to discover gene signatures for EAOC.
(A) Workflow of the gene set regularity model. The gene set regularity (GSR) index was computed by converting the gene expression ordering of gene elements in a gene set through the Gene Ontology (GO) term or canonical pathway databases. The informativeness of the GSR index was assessed by the accuracy of recognition, classification, and prediction by machine learning using binary or multiclass classifications. Functionome analyses were carried out to investigate the pathogenesis of endometriosis (ES), clear cell carcinoma (CCC), endometrioid ca (EC) and endometriosis-associated ovarian carcinoma (EAOC) by statistical methods, hierarchical clustering, and exploratory factor analysis. (B) Heatmaps and dendrogram for the three diseases. The dendrogram (left side of the heatmap) showed the relationship of the three diseases. When displayed on the heatmap, each of the three diseases computed through either the GO term gene sets showed a distinct pattern. However, the patterns were more similar between CCC and EC.