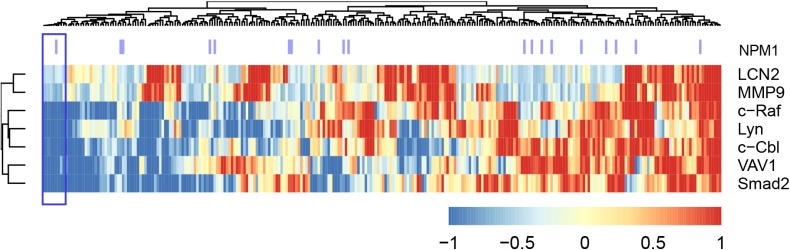
Figure 9. Clustering analysis of the TARGET RNA-Seq data set, using the gene expression data for the genes we analyzed by western blot (Figure 8).
Gene expression data points were first z-normalized and then the distance between points was measured using Euclidean distance and then the complete agglomeration method was used in clustering by the ‘hclust’ function in R package ‘stats’. The heatmap using the ‘aheatmap’ function in R package ‘NMF’ (https://github.com/renozao/NMF). Upregulation is color coded red and downregulation is color coded blue.