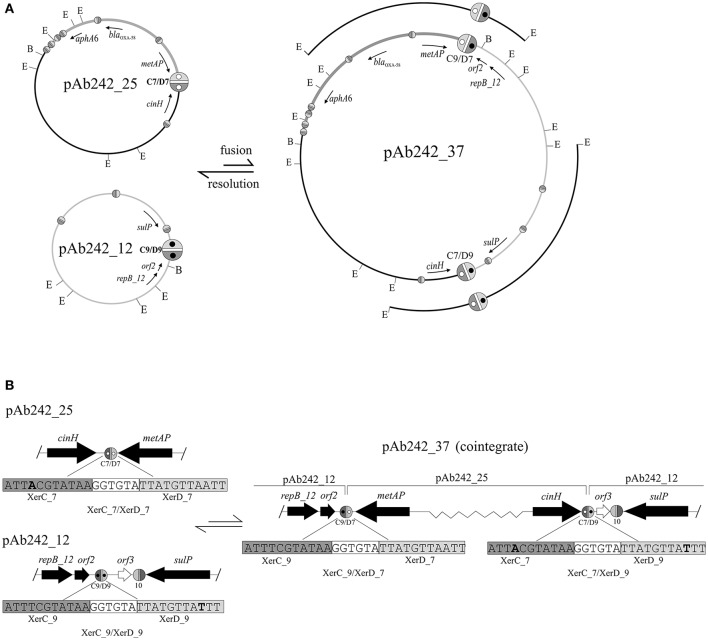
Figure 3.
Site-specific recombination between XerC/D sites mediates fusion between pAb242_25 and pAb242_12. (A, Upper panel) Scheme depicting the formation of the plasmid co-integrate pAb242_37 by inter-molecular site-specific recombination at the sister pair XerC_7/XerD_7 (C7/D7) and XerC_9/XerD_9 (C9/D9) located in pAb242_25 and pAb242_12, respectively. The resolution of pAb242_37 by intra-molecular site-specific recombination at the new pair of XerC/D sites generated during the fusion reaction (C9/D7 and C7/D9, respectively) is also shown. The blaOXA-58- and aphA6-containing adaptive module in pAb242_25 and pAb242_37 is highlighted in dark gray. The positions of EcoRI (E) and BamHI (B) restriction sites in these plasmids are also indicated. The different XerC/D-like recognition sites (see Table 2 for sequence details) are highlighted as ovals (not drawn to scale) as detailed in the legend to Figure 2. The pair of sites identified as recombinationally active in this work have been arbitrarily enlarged. Open and closed inner circles within XerC_7/XerD_7 and XerC_9/XerD_9, respectively, are shown to facilitate the visualization of the fusions generating XerC_9/XerD_7 (C9/D7) and XerC_7/XerD_9 (C7/D9) in pAb242_37. The cloned EcoRI fragments used to identify these fusions (see text for details) are also indicated above the pAb242_37 scheme. (B, Lower panel) Enlarged vision including the nucleotide sequences of the XerC/D sites involved in the fusion/resolution reactions showing that the polarity of the sites was maintained after site-specific recombination. Relevant nucleotides in each site are highlighted in bold. Slash (/) symbols at the borders indicate that further sequence data existed beyond this position. The scheme is not drawn to scale.