ABSTRACT
The fifth wave of A(H7N9) virus infection in China from 2016 to 2017 caused great concern due to the large number of individuals infected, the isolation of drug-resistant viruses, and the emergence of highly pathogenic strains. Antibodies against neuraminidase (NA) provide added benefit to hemagglutinin-specific immunity and may be important contributors to the effectiveness of A(H7N9) vaccines. We generated a panel of mouse monoclonal antibodies (MAbs) to identify antigenic domains on NA of the novel A(H7N9) virus and compared their functional properties. The loop formed in the region of residue 250 (250 loop) and the domain formed by the loops containing residues 370, 400, and 430 were identified as major antigenic regions. MAbs 1E8, 2F6, 10F4, and 11B2, which recognize these two antigenic domains, were characterized in depth. These four MAbs differ in their abilities to inhibit cleavage of small and large substrates (methyl-umbelliferyl-acetyl neuraminic acid [MU-NANA] and fetuin, respectively) in NA inhibition assays. 1E8 and 11B2 did not inhibit NA cleavage of either MU-NANA or fetuin, and 2F6 inhibited cleavage of fetuin alone, whereas 10F4 inhibited cleavage of both substrates. All four MAbs reduced the in vitro spread of viruses carrying either the wild-type N9 or N9 with antiviral-resistant mutations but to different degrees. These MAbs have different in vivo levels of effectiveness: 10F4 was the most effective in protecting mice against challenge with A(H7N9) virus, 2F6 was less effective, and 11B2 failed to protect BALB/c mice at the doses tested. Our study confirms that NA-specific antibodies can protect against A(H7N9) infection and suggests that in vitro properties can be used to rank antibodies with therapeutic potential.
IMPORTANCE The novel A(H7N9) viruses that emerged in China in 2013 continue to infect humans, with a high fatality rate. The most recent outbreak resulted in a larger number of human cases than previous epidemic waves. Due to the absence of a licensed vaccine and the emergence of drug-resistant viruses, there is a need to develop alternative approaches to prevent or treat A(H7N9) infection. We have made a panel of mouse monoclonal antibodies (MAbs) specific for neuraminidase (NA) of A(H7N9) viruses; some of these MAbs are effective in inhibiting viruses that are resistant to antivirals used to treat A(H7N9) patients. Binding avidity, inhibition of NA activity, and plaque formation correlated with the effectiveness of these MAbs to protect mice against lethal A(H7N9) virus challenge. This study identifies in vitro measures that can be used to predict the in vivo efficacy of NA-specific antibodies, providing a way to select MAbs for further therapeutic development.
KEYWORDS: A(H7N9), influenza, neuraminidase, monoclonal antibody, antiviral
INTRODUCTION
The novel A(H7N9) influenza virus that emerged in China in 2013 (1) continues to cause infections in humans, with approximately 40% mortality (2, 3). According to the Food and Agriculture Organization (FAO) website, as of 25 October 2017, 1,622 laboratory-confirmed A(H7N9) cases have been reported, 619 of which have been fatal. While there is no evidence of sustained human-to-human transmission to date, this virus has properties that suggest that it could easily become adapted to replication in humans. Candidate vaccine viruses (CVVs) of A(H7N9) have been generated and tested for safety and immunogenicity (4–6) and could be authorized for use if needed. The most recent outbreak of A(H7N9) infection, i.e., the fifth epidemic wave, has increased the concern regarding the potential pandemic threat of this virus because the incidence rate has increased, A(H7N9) strains with hemagglutinins (HAs) that are antigenically distinct from the tested CVVs have emerged, and highly pathogenic strains have been isolated (7, 8). There is therefore a need to consider additional ways to prevent and control A(H7N9) infection.
Neuraminidase (NA) plays a critical role in the replication and spread of influenza virus (7, 9). Antibodies that inhibit NA activity correlate with reduced clinical signs of influenza and shortened duration of virus replication (10). Although current influenza vaccines do not contain a standard amount of NA, there are many examples demonstrating increased NA inhibition (NI) antibody titers following vaccination (11, 12) and a correlation between NI titers and vaccine effectiveness (13–16). While a recombinant HA-based A(H7N9) vaccine has been developed and clinically evaluated (4), the contribution of NA immunity against A(H7N9) infection has been less explored. The antigenic structure of N9 has been described in studies of A/tern/Australia/G70C/75 (G70C, H11N9) and A/whale/1/84 (H13N9), with X-ray crystallography of N9 and monoclonal antibody (MAb) complexes defining epitopes that surround the enzyme active site (17–20). However, the antigenic characteristics of the NA of recent A(H7N9) viruses are poorly defined. A recent report showed that amino acids around the enzyme active center are critical for binding of an N9 MAb, 3c10-3 (21). In the present study, we characterize a panel of MAbs that bind to the NA of a reference A(H7N9) virus, A/Anhui/1/2013 (AH/13), and evaluate the in vitro functional attributes and in vivo effectiveness of selected N9 MAbs that bind to different antigenic domains.
RESULTS
Preparation of N9 MAbs.
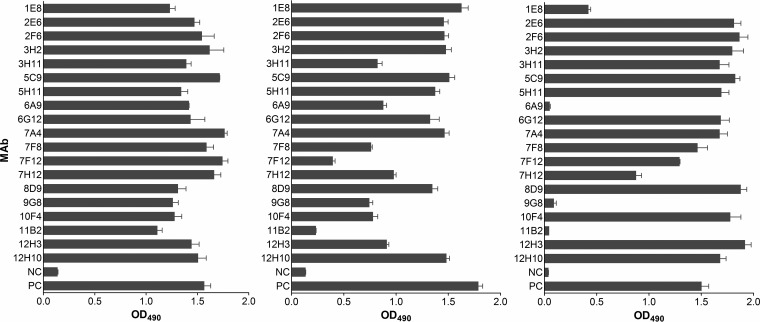
A panel of 19 MAbs against the NA of the novel A(H7N9) virus AH/13 was generated through routine mouse hybridoma technology (22, 23). All of these antibodies are of IgG1 isotype. The specificity of each antibody for N9 was confirmed by cell-based enzyme-linked immunosorbent assay (ELISA), in which the binding of the MAb was tested with NA transiently expressed in 293T human embryonic kidney cells (23). As shown in Fig. 1, all MAbs bound the NA of AH/13, and the majority also reacted with the NA of the H11N9 virus G70C and a recent A(H7N9) reference virus, A/Guangdong/17SF0003/2016 (GD/16). MAb 3A2, which is specific for the NA of a seasonal H1N1 virus, A/Brisbane/59/2007 (23), was used throughout our study as a negative control.
FIG 1.
Binding of N9 MAbs to the NA of AH/13, G70C, and GD/16. Binding was measured in cell-based ELISAs using AH/13 NA (left panel) and G70C NA (center panel) transiently expressed on 293T cells as well as purified H6N9GD/16 virus (right panel). Cell culture supernatant of each hybridoma was tested irrespective of the actual antibody concentration. NC, negative control, cell culture supernatant of N1-specific MAb 3A2; PC, positive control, mouse hyperimmune serum against AH/13 NA (left and right panels) and G70C NA (middle panel). Shown are mean OD490 values plus standard deviations of two independent assays run in duplicate wells.
Based on the ELISA binding profile (Fig. 1) and early measurements to assess NI activity of hybridoma cell culture supernatants (data not shown), the following 10 MAbs were selected for further characterization in various in vitro assays: 1E8, 2E6, 2F6, 5C9, 5H11, 7A4, 7F8, 7F12, 10F4, and 11B2. We sought to perform the in vitro assays using the A(H7N9) CVV CBER-RG4A; however, the plaques formed by this virus in Madin-Darby canine kidney (MDCK) cells were too small (data not shown) to evaluate the impact of MAbs on virus spread in cells. Therefore, a reassortant H6N9AH/13 virus, which bears the HA of A/turkey/Massachusetts/3740/1965 (H6N2) and the NA of AH/13 was used in in vitro assays.
N9 residues critical for MAb binding.
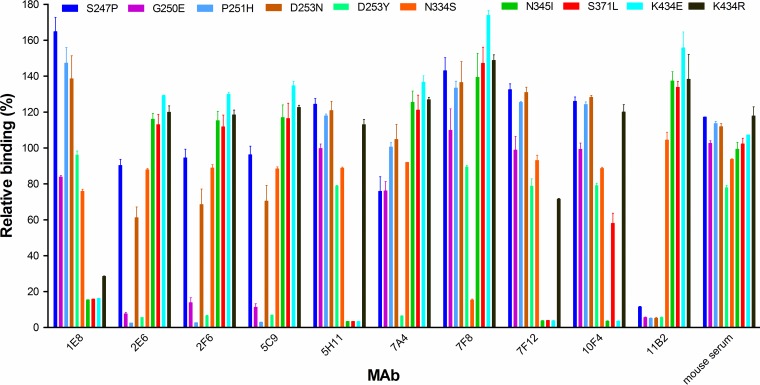
To localize residues on N9 that are critical for antibody binding, H6N9AH/13 virus escape mutants were selected in eggs and plaque purified in MDCK cells as previously described (23). The NA of each mutant virus was sequenced and aligned with that of the parent virus. Ten mutations at 8 amino acid positions, i.e., S247P, G250E, P251H, D253N, D253Y, N334S, N345I, S371L, and K434E or K434R [G70C NA numbering was followed; the NA of the novel A(H7N9) viruses, including AH/13, has 5 fewer amino acids], were identified in the NA of escape mutants selected by the 10 antibodies (Table 1). Each mutant contains only a single amino acid mutation in NA although some MAbs, i.e., 1E8, 5C9, 5H11, and 7F12, selected more than one escape mutant. We measured the binding of each selecting MAb to the 10 escape mutants by ELISA. As shown in Fig. 2, the selecting MAbs either completely lost binding or had substantially reduced binding to the selected mutant viruses. MAb binding was often also impacted by mutations that were not selected by the MAb itself but, rather, by other MAbs. For instance, MAb 11B2 selected the S247P mutation in the NA, but mutations at residues 250, 251, and 253, which were selected by 2E6 and 5C9, also abolished binding. MAb 1E8 selected mutations at residue 434, but mutations at residues 345 and 371, selected by MAbs 5H11 and 7F12, respectively, also impacted the binding of MAb 1E8 to NA. To ensure that the variants were not selected nonspecifically during passaging, the escape mutant selection was also performed with the control antibody 3A2; none of the viruses sequenced from these selections contained mutations in NA.
TABLE 1.
Purified N9 MAbs for in vitro and in vivo studies
| MAb | IC50 (ng/ml)a |
Mutation in NA of selected mutants | KD (nM)b | |
|---|---|---|---|---|
| ELLA | MU-NANA | |||
| 1E8c | >4,000 | >32,000 | K434E or K434R | >335d |
| 2E6 | 35.5 | >32,000 | P251H | 2.4 |
| 2F6 | 20.6 | >32,000 | D253N | 3.7 |
| 5C9 | 26.5 | >32,000 | G250E or P251H | 93.2 |
| 5H11 | 142.9 | >32,000 | N345I or K434E | >335 |
| 7A4 | 161.2 | >32,000 | D253Y | 16.8 |
| 7F8 | >4,000 | >32,000 | N334S | 46.9 |
| 7F12 | 305.5 | >32,000 | S371L or K434E | >335 |
| 10F4 | 47.6 | 643 | K434E | 1.4 |
| 11B2 | >4,000 | >32,000 | S247P | 250 |
ELLAs and MU-NANA assays were performed with reassortant H6N9AH/13, which bears the HA gene of the H6N2 virus A/turkey/Massachusetts/3740/1965, the NA of AH/13, and the internal genes of the A(H1N1) virus A/Puerto Rico/8/1934 (PR8).
KD values were measured with SPR using recombinant AH/13 NA.
MAbs 1E8, 2F6, 10F4, and 11B2 were selected for in vivo experiments.
No binding was detected by SPR at the highest MAb concentration tested, 50 μg/ml (335 nM).
FIG 2.
Binding of N9 MAbs to escape mutant viruses. Binding was measured in ELISAs using purified H6N9AH/13 parent and escape mutants. The MAbs were tested at 1 μg/ml. The OD490 values were normalized to the signals obtained with the parent H6N9AH/13 and expressed as relative binding. Shown are the mean OD490 values plus standard deviations from two independent assays run in duplicate wells.
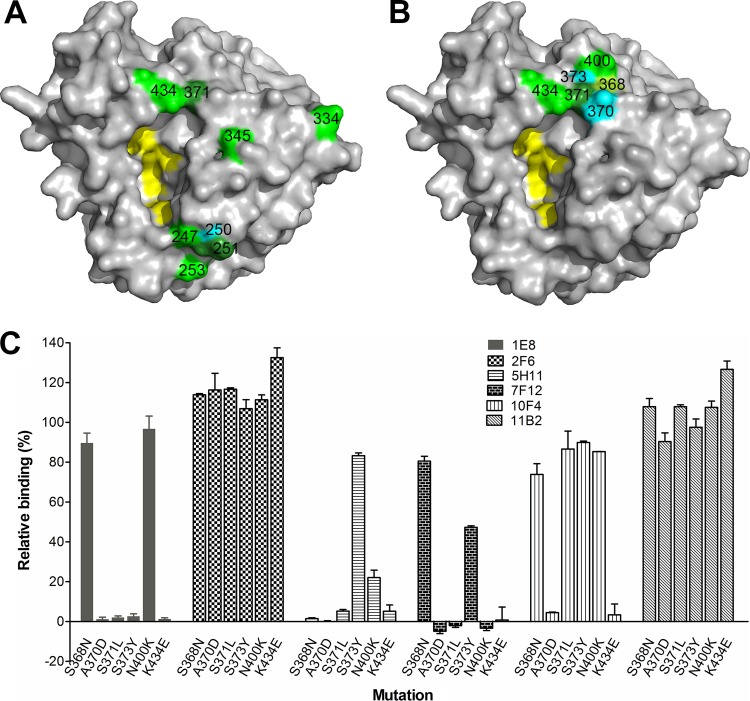
The residues that impact MAb binding are located on the top surface of N9 and mostly in close proximity to the enzyme active site (Fig. 3A). Amino acid 371, selected by MAb 7F12, and residue 434, selected by MAbs 1E8, 5H11, 7F12, and 10F4, are either within or close to loops containing residues 370, 400, and 430 that give N9 the ability to agglutinate red blood cells. This hemabsorbing (HB) site on N9, which is independent of the enzyme active site (24), was first identified in the NA of G70C (25) and is also present in the NA of AH/13 (26). To further investigate the specificity of MAbs 1E8, 5H11, 7F12, and 10F4 that bind residues in the HB region, single amino acid mutations were introduced into the AH/13 NA gene at positions 368, 370, 371, 373, 400, and 434 (Fig. 3B) in the pCAGGS expression plasmid. 293T cells were transfected with mutant and wild-type (wt) plasmids and used in cell-based ELISAs to measure MAb binding to the transiently expressed NAs. MAbs 2F6 and 11B2, which recognize residues in the 250 loop (residues 253 and 247, respectively), were included in the assay for comparison. As shown in Fig. 3C, each MAb was impacted differently by the mutations. Mutations at residues 370, 371, 373, and 434 abolished the binding of NA by MAb 1E8, whereas mutations at residues 368 and 400 had little impact. 5H11, 7F12, and 10F4 had reduced binding to all mutations compared to that of the wt NA; however, the degrees of binding differed. Binding of MAb 5H11 was either abolished or significantly reduced (∼20% relative binding compared to that of wt AH/13 NA) when mutations were introduced at positions 368, 370, 371, 400, and 434. This antibody retained >80% binding to the S373Y mutant. In contrast, the binding of antibody 7F12 to NA was dramatically reduced by mutations at residues at 370, 371, 373, 400, and 434 but not by mutation at residue 368. Mutation at position 370 or 434 abolished the binding of 10F4 to NA, while changes at the other amino acids in loops associated with the HB site had much less impact on 10F4 binding. The binding of antibodies 2F6 and 11B2 was not reduced compared to that of the wt NA. These results confirm that the epitopes recognized by MAbs 1E8, 5H11, 7F12, and 10F4 include amino acids within the three loops that constitute the HB site and indicate that the fine specificity of each antibody is unique.
FIG 3.
Residues on N9 that are critical for MAb binding. (A) Locations of the eight key residues (identified through escape mutant selection) on an N9 monomer (PDB accession number 4MWW). The image was generated with PyMOL software (Delano Scientific). Different colors were used to differentiate neighboring residues when necessary. (B) Residues in the 370, 400, and 430 loops that constitute the HB site on N9 that endow it with its ability to agglutinate red blood cells. Residues 118, 119, 224, 227, and 406 are highlighted in yellow in both panels A and B to depict the location of the NA active center. (C) Impact of amino acid mutations within the HB site of AH/13 NA on the binding by MAbs 1E8, 2F6, 5H11, 7F12, 10F4, and 11B2. Binding was measured by cell-based ELISA, with the signals expressed as percentages of those obtained with wt AH/13 NA. Shown are the means plus standard deviations from two independent assays run in duplicate wells.
Taking these observations together, 12 antigenically critical amino acid positions, i.e., 247, 250, 251, 253, 334, 345, 368, 370, 371, 373, 400, and 434, on AH/13 NA were identified. To examine the variation at these NA positions in more recent A(H7N9) virus isolates, 169 full-length NA sequences of human A(H7N9) viruses isolated during the fifth epidemic wave, which are available from the Global Initiative on Sharing All Influenza Data (GISAID [http://www.gisaid.org]), were compared to the AH/13 NA sequence at these 12 positions. As shown in Table 2, the majority (94.7%) of viruses of the fifth epidemic wave bear a different residue from that of AH/13 at position 247. Amino acid variations at positions 334, 345, and 370 were also observed. At the remaining eight critical amino acid positions, all of the isolates of the fifth epidemic wave possess residues that are identical to those in AH/13 NA.
TABLE 2.
Amino acids at NA residues of A(H7N9) viruses that are critical for MAb binding
| Residue no.a | Amino acids located in NA of: |
|
|---|---|---|
| AH/13 | Viruses isolated in 2016–2017b | |
| 247 | S | S (4.7), P (94.7), P/L (0.6)c |
| 250 | G | G (100) |
| 251 | P | P (100) |
| 253 | D | D (100) |
| 334 | N | N (99.4), D (0.6) |
| 345 | N | N (97), I (2.4), N/I (0.6) |
| 368 | S | S (100) |
| 370 | A | A (89.3), T (10.1), I (0.6) |
| 371 | S | S (100) |
| 373 | S | S (100) |
| 400 | N | N (100) |
| 434 | K | K (100) |
N9 numbering is based on the G70C NA.
A total of 169 full-length NA sequences of human A(H7N9) viruses isolated between 1 October 2016 and 20 October 2017 (GISAID) were compared with the sequence of AH/13 at 12 antigenically critical residues. Numbers in parentheses represent the percentages of A(H7N9) isolates bearing each residue in NA.
A mixed sequence was present at positions 247 and 345.
Inhibition of NA activity by N9 MAbs.
The 10 selected MAbs were tested in fluorescence or enzyme-linked lectin inhibition assays (ELLAs) that use a small (methyl-umbelliferyl-acetyl neuraminic acid [MU-NANA], 489 Da) or large (fetuin, 49,000 Da) substrate, respectively. H6N9AH/13 virus was used as the antigen in both assays. Most of the MAbs inhibited the ability of N9 to cleave fetuin; however, the 50% inhibition concentrations (IC50s) differed. Some antibodies inhibited N9 activity very efficiently in this assay (ELLA), with IC50s of <50 ng/ml, while others required higher concentrations to inhibit NA, with IC50s between 100 and 300 ng/ml (Table 1). Three MAbs, 1E8, 7F8, and 11B2, did not inhibit NA activity in either assay. In the assay using the MU-NANA substrate, the only MAb that inhibited cleavage was 10F4, with an IC50 of 643 ng/ml, while the remaining nine antibodies failed to inhibit NA activity at the highest tested concentration, 32 μg/ml (Table 1).
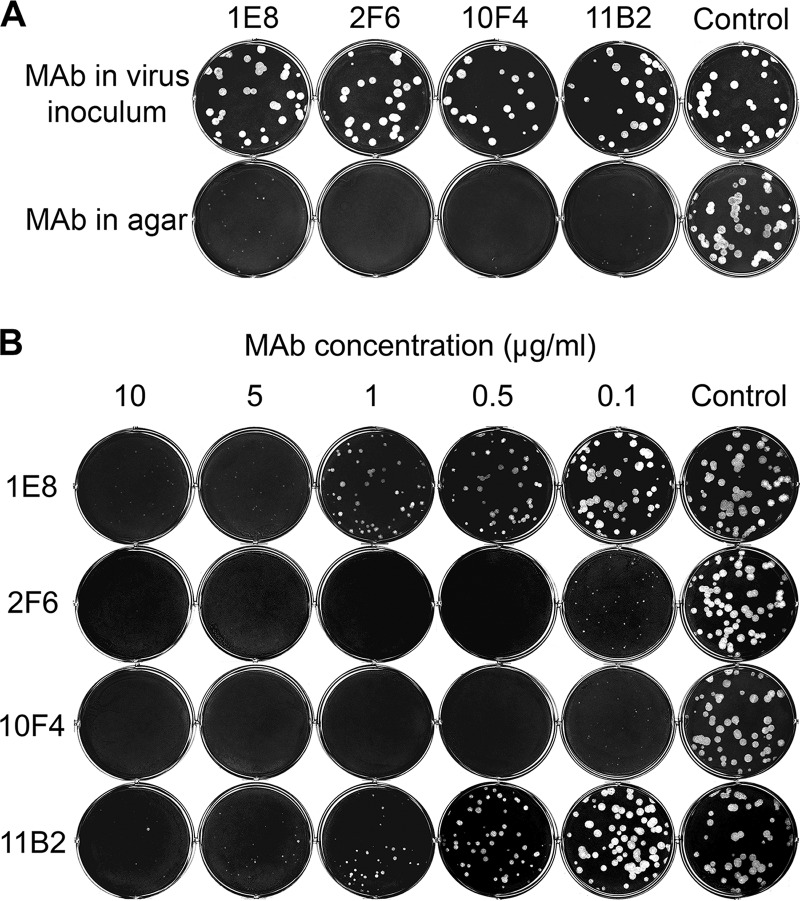
We next examined the ability of MAbs 1E8, 2F6, 10F4, and 11B2 to inhibit plaque formation by virus in MDCK cells, with each MAb supplemented into the agar overlay. These four antibodies were tested because they inhibited cleavage of small and large substrates by NA differently, and based on the critical residues identified (Table 1; Fig. 2 and 3C), they likely bind to different antigenic domains. In addition, they have different binding affinities to NA, as indicated by the equilibrium dissociation constants (KDs) (Table 1) measured with surface plasmon resonance (SPR) (27, 28). 10F4 and 2F6 have higher avidity for NA (KD values of 1.4 and 3.7 nM, respectively) than 1E8 and 11B2 (no binding was detected by SPR at the highest antibody concentration tested, 335 nM). None of these MAbs inhibited the formation of H6N9AH/13 plaques when they were applied in the virus inoculum before infection of cells (Fig. 4A), suggesting that these antibodies did not block the initial entry of virus into cells; but when these antibodies were supplemented in the agar overlay after H6N9AH/13 infection, they substantially inhibited plaque formation in a dose-dependent manner (Fig. 4A, lower panel, and B). 2F6 and 10F4 reduced plaque size when applied at 0.1 μg/ml in the agar overlay, while ≥5 μg/ml of 1E8 and 11B2 was needed to achieve similar inhibition. At a concentration of ≥0.5 μg/ml, both 2F6 and 10F4 prevented plaque formation, whereas small plaques were still observed at the highest 1E8 and 11B2 concentration tested (10 μg/ml).
FIG 4.
N9 MAbs inhibit H6N9AH/13 plaque formation. MAbs 1E8, 2F6, 10F4, and 11B2 did not affect plaque formation when supplemented in the virus inoculum but inhibited plaque formation when supplemented in agar overlay after cells were infected with virus. (A) MDCK cells growing in six-well plates were infected with ∼30 to 40 PFU of H6N9AH/13 premixed with each MAb at 5 μg/ml and covered with agar without antibody after thorough washing (upper panel) or infected with H6N9AH/13 and then covered with agar containing antibody (5 μg/ml) after washing (lower panel). Control, N1-specific MAb 3A2. (B) MDCK cells were infected with H6N9AH/13 and then overlaid with agar that contained various concentrations of MAbs 1E8, 2F6, 10F4, and 11B2 (0.1 to 10 μg/ml) or the control MAb, 3A2 (10 μg/ml).
N9 MAbs block the release of progeny virions.
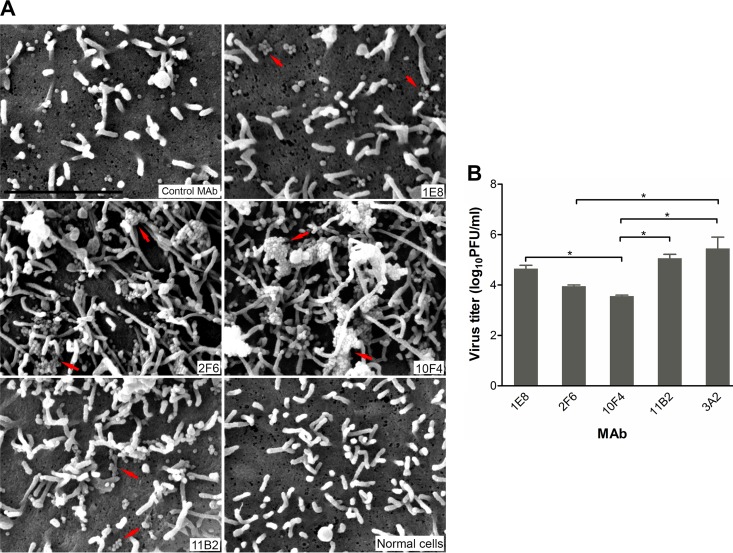
The reduction in virus plaque size by MAbs 1E8 and 11B2 was somewhat surprising since they had not inhibited enzyme activity in NI assays. While this can be explained by the different assay format and input virus load, we wanted to demonstrate that these two MAbs as well as those (2F6 and 10F4) that inhibit NA activity had a direct impact on NA function. MDCK cells growing on coverslips were infected with H6N9AH/13 and cultured for 10 h in medium supplemented with each MAb (10 μg/ml) and then processed and viewed under a scanning electron microscope (SEM). Images in Fig. 5 show that virus particles, mostly spherical, were evident on infected cells (all panels of Fig. 5A, except the bottom right panel, which is an image of uninfected control cells). In contrast to cells incubated with the control antibody 3A2 shown in the top left panel, which has a few dispersed single virus particles, aggregates of progeny virus particles were observed on the surface of cells cultured in the presence of N9-specific MAbs. While the aggregates were relatively small, involving a few virus particles in the presence of antibodies 1E8 and 11B2, many bigger virus aggregates were observed on the surface of cells treated with either 2F6 or 10F4. Consistent with this observation, virus titers in supernatants of cells treated with 1E8 and 11B2 were only slightly lower than the titer in supernatants from cells treated with the control antibody 3A2, while 2F6 and 10F4 resulted in virus titers that were 30- and 100-fold lower, respectively (P < 0.05) (Fig. 5B). These findings provide direct evidence that N9 MAbs inhibit virus spread by blocking the release of progeny virions from the infected cells, with 2F6 and 10F4 being more effective than 1E8 and 11B2.
FIG 5.
N9 MAbs block virus release from infected cells. MDCK cells were infected with H6N9AH/13, and MAbs (N9 MAbs 1E8, 2F6, 10F4, and 11B2 and the control MAb 3A2) were supplemented at 10 μg/ml in cell culture medium after infection. Cells were fixed 10 h after virus infection for SEM observation, and supernatants were collected for virus titration. (A) SEM images showing virus particles/aggregates on the cell surface. Arrows point to virus aggregates. Bar, 2 μm. (B) Virus titers in cell culture medium were measured by plaque assay. Data are the means plus standard deviations of two independent experiments. *, P < 0.05.
N9 MAbs are effective against drug-resistant viruses.
Because the novel A(H7N9) virus is resistant to M2 blockers, oseltamivir, an NA inhibitor, is commonly used to treat patients with A(H7N9) infection (1, 29, 30). A number of oseltamivir-resistant clinical A(H7N9) isolates have been identified. Mutations in the NA that are associated with antiviral resistance in these viruses include E119V, I222R or I222K, A246T, and R292K (8, 29, 31, 32). We rescued H6N9AH/13 viruses that contained 119V, 222R, 246T, or 292K in the NA and tested the ability of MAbs 1E8, 2F6, 11B2, and 10F4 to inhibit their NA activity. Reassortant viruses containing these drug-resistant mutations had significantly reduced (P < 0.05 at virus dilutions of ≤256) enzyme activity compared to that of the parent H6N9AH/13 in an MU-NANA assay. The NA activity of H6N9AH/13 292K was hardly detectable when the virus was incubated with MU-NANA for 1 h, the usual length of incubation with MU-NANA (Fig. 6A). Extending the incubation time to 3.5 h increased the catalysis of substrate, but the virus was still less effective than other mutants or virus with wt N9. Using a more acidic buffer (pH 5.3) dramatically enhanced the NA activity of H6N9AH/13 292K virus (Fig. 6A), allowing the sensitivity of each reassortant virus to NA inhibitors to be measured. An MU-NANA assay was consequently conducted under optimal pH conditions, i.e., pH 5.3 buffer for H6N9AH/13 292K and pH 7.2 buffer for other viruses. As shown in Table 3, the mutant NAs are resistant to both oseltamivir and zanamivir, as indicated by increased IC50 measurements compared to values for the wt N9.
FIG 6.
N9 MAbs inhibit the NAs of viruses resistant to antivirals. (A) NA activity of H6N9AH/13 viruses with antiviral-resistant mutations E119V, I222R, A246T, and R292K in the NA, measured by MU-NANA assay with viruses incubated with the substrate for 1 h (left panel) and 3.5 h (center panel) or with a low-pH (5.3) buffer (right panel). RFU, relative fluorescence units. Data represent the means plus standard deviations of two independent assays run in duplicate. (B) Plaques of H6N9AH/13 mutant viruses in MDCK cells cultured at 33°C (upper panel) and 37°C (lower panel). (C) Plaque sizes of the H6N9AH/13 mutant viruses described in panel B. *, P < 0.05. (D) Plaques of H6N9AH/13 mutant viruses in MDCK cells cultured in the presence of N9 MAbs 1E8, 2F6, 10F4, and 11B2 and the control MAb 3A2 in the agar overlay. 1E8, 11B2, and 3A2 were supplemented at 5 μg/ml, whereas 2F6 and 10F4 were supplemented at 1 μg/ml.
TABLE 3.
Susceptibility of H6N9AH/13 mutant viruses to NA inhibitors
| Virusa | Oseltamivir |
Zanamivir |
||
|---|---|---|---|---|
| IC50 (nM)b | Fold change in IC50c | IC50 (nM) | Fold change in IC50 | |
| wt H6N9AH/13 | 0.55 ± 0.06 | 2.31 ± 0.12 | ||
| H6N9AH/13 119V | 35.13 ± 1.03 | 64 | 11.79 ± 0.29 | 5 |
| H6N9AH/13 222R | 29.62 ± 3.19 | 54 | 24.11 ± 2.01 | 10 |
| H6N9AH/13 246T | 1.70 ± 1.00 | 3 | 37.00 ± 1.27 | 16 |
| H6N9AH/13 292K | >200 | >363 | 123.26 ± 8.13 | 53 |
H6N9AH/13 viruses were rescued to introduce each of the single antiviral-resistant mutations, E119V, I222R, A246T, and R292K, in NA.
IC50 denotes the concentration of NA inhibitors (oseltamivir and zanamivir) that reduces NA activity by 50% in the MU-NANA assay. Shown are means ± standard deviations from two independent assays run in duplicate wells.
Fold change relative to the wt level.
Since MAbs 1E8 and 11B2 did not inhibit the activity of wt NA in NI assays (Table 1), the sensitivity of the mutant NAs to inhibition by MAbs was tested in plaque assays. H6N9AH/13 119V, H6N9AH/13 222R, and H6N9AH/13 246T formed plaques with sizes comparable to those of the parent H6N9AH/13, while H6N9AH/13 292K formed significantly smaller (P < 0.05) plaques at both 33°C and 37°C (Fig. 6B and C). Because H6N9AH/13 292K plaques could be more accurately measured when the assay was performed at 37°C, the plaque assay with this virus was conducted by incubating the virus infected-cells at 37°C, while the cells for assays with the other three mutant viruses were incubated at 33°C. As shown in Fig. 6D, antibodies 1E8, 2F6, 10F4, and 11B2 inhibited plaque formation by the drug-resistant mutants as effectively as they did with virus bearing wt N9.
N9 MAbs differ in protective effectiveness against lethal A(H7N9) virus challenge.
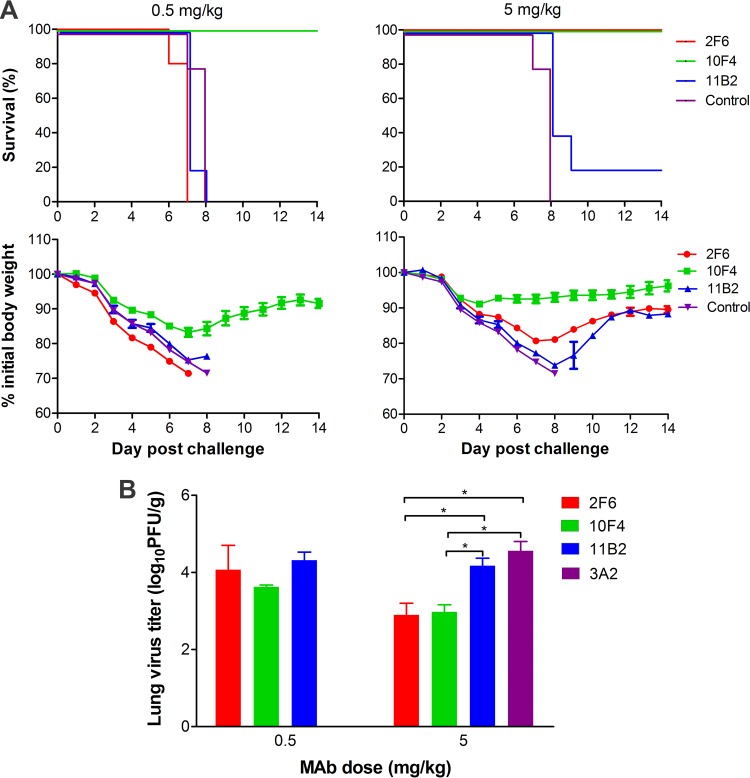
Mouse studies to assess the protective effectiveness of N9 MAbs against wt A(H7N9) virus challenge were conducted with MAbs 2F6, 10F4, and 11B2. Antibodies were administered to BALB/c mice intraperitoneally (i.p.) at either 0.5 or 5 mg/kg 12 h before intranasal (i.n.) challenge with 10 median mouse lethal doses (MLD50) of wt AH/13 virus (Fig. 7A). Treatment with MAb 11B2 did not protect any mice from death at 0.5 mg/kg, and only 1 out of the 5 mice treated with 5 mg/kg survived the virus challenge. The 0.5-mg/kg dose of 2F6 failed to protect mice from death, but all mice were protected by the 5-mg/kg dose although the mice experienced weight loss. MAb 10F4 was the most effective treatment and prevented mortality at both 0.5 and 5 mg/kg. From day 5 postchallenge (p.c.) onwards, weight loss in the 10F4 group was significantly less than that of all other groups (P < 0.05).
FIG 7.
N9 MAbs vary in protecting mice from lethal A(H7N9) virus challenge. BALB/c mice (n = 10 per group) were treated i.p. with MAbs at 0.5 or 5 mg/kg, followed by i.n. challenge with 10 MLD50 of wt AH/13 12 h later. The N1-specific MAb 3A2 was administered at 5 mg/kg as a negative control. Survival and weight loss (n = 5 per group) were monitored for 14 days. Lungs from another five mice were collected on day 6 p.c. for virus titration (n = 3) and histological examination (n = 2) (Fig. 8). (A) Survival and weight loss. (B) Lung virus titers expressed as log10 PFU/g. Data are the means plus standard deviations (n = 3). *, P < 0.05.
Virus titers in the lungs collected from mice (n = 3) on day 6 p.c. were measured by plaque assay. Treatment with 11B2 at doses of 0.5 and 5 mg/kg and 2F6 at 0.5 mg/kg did not significantly reduce virus load compared to the result with the control antibody 3A2, which was administered only at 5 mg/kg; in contrast, at a dose of 5 mg/kg, virus titers of the 2F6 and 10F4 groups were significantly lower than the titer of the control (∼40-fold lower; P < 0.05). The lung virus titers of mice treated with 10F4 at 0.5 mg/kg were also significantly lower than the titer of the control (∼10-fold lower; P < 0.05) (Fig. 7B).
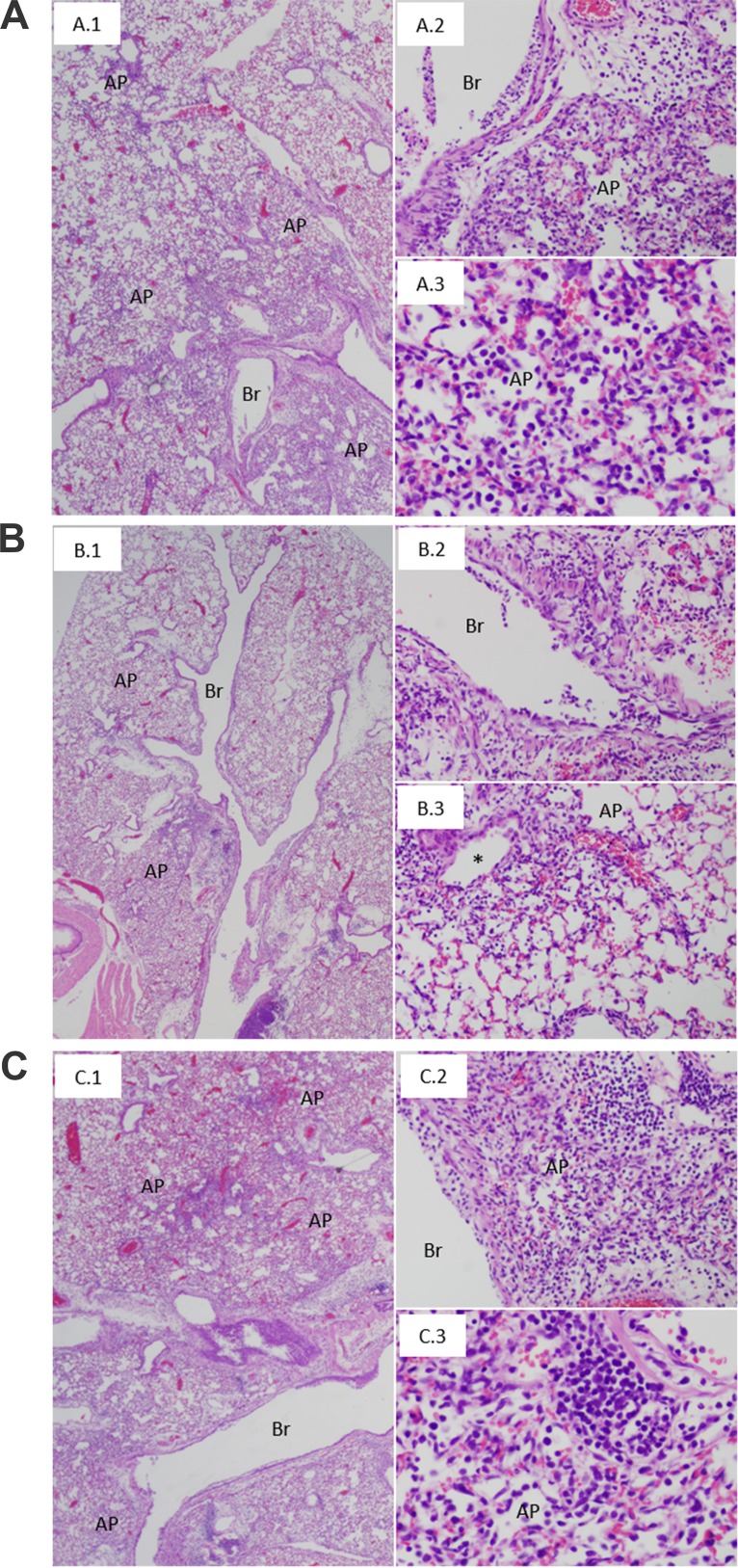
Histopathological analyses of lungs from the infected mice were conducted blindly by a pathologist on hematoxylin and eosin (H&E)-stained sections. Entire lung cross sections from the two mice per treatment group were evaluated. AH/13-infected mice treated with the control MAb 3A2 showed widespread lesions, including necrotizing bronchitis and bronchiolitis, with transmural infiltration of inflammatory cells and respiratory epithelial cell necrosis (Fig. 8A, frame 1). The lungs showed an acute pneumonia, affecting approximately 40 to 50% of the lung parenchyma, consisting of interstitial inflammatory infiltrates and acute alveolitis (Fig. 8A, frame 2) with a mixed inflammatory cell infiltrate, including prominent neutrophils (Fig. 8A, frame 3). Lungs from AH/13-infected mice treated with N9 MAbs 2F6, 10F4, and 11B2 at the 0.5-mg/kg dose had pathology similar to that of the MAb 3A2-treated control animals (data not shown); however, lungs from mice treated with 2F6 and 10F4 at the 5-mg/kg dose had significantly less pathology (Fig. 8B). In these animals, widespread bronchitis and bronchiolitis were similar to those observed in control animals, but acute pneumonia lesions were observed only in approximately 20% of the lung parenchyma (Fig. 8B), usually involving alveoli in peribronchiolar regions (Fig. 8B, frame 3). Based on blinded observations of complete lung sections, the proportion of lung containing lesions was observably less in the 2F6 and 10F4 groups than in the 3A2-treated control group. MAb 11B2-treated animals had histopathological lesions indistinguishable from those of MAb 3A2-treated animals, with widespread bronchitis, bronchiolitis, and acute pneumonia involving 40 to 50% of the lung parenchyma (Fig. 8C).
FIG 8.
Representative lung histopathological lesions from mice treated with N9 MAbs and challenged with A(H7N9) virus. The antibody treatment (5 mg/kg) and virus challenge were as described in the legend of Fig. 7. (A) Tissues from mice treated with the control MAb 3A2 show the following: widespread bronchitis (Br), bronchiolitis, and acute pneumonia (AP) affecting approximately 50% of the lung parenchyma (A.1); necrotizing bronchitis with transmural infiltration and acute pneumonia consisting of a prominent interstitial and intra-alveolar inflammatory infiltrate (A.2); acute pneumonia consisting of a mixed inflammatory infiltrate with prominent neutrophils (A.3). (B) Tissues from mice treated with N9 MAb 10F4 show the following: bronchitis, bronchiolitis, and acute pneumonia affecting approximately 20% of the lung parenchyma (B.1); necrotizing bronchitis with transmural infiltration (B.2); focus of acute pneumonia surrounding a bronchiole (*), consisting of a mixed inflammatory infiltrate (B.3). (C) Tissues from mice treated with N9 MAb 11B2 show lung lesions (C.1, C.2, and C.3) similar to those observed in mice treated with the control MAb (A). Magnifications, ×20 (A.1, B.1, and C.1), ×200 (A.2, B.2, B.3, and C.2), and ×400 (A.3 and C.3).
DISCUSSION
This study characterizes MAbs that bind different antigenic domains within the NA of A(H7N9) viruses and compares their in vitro functional attributes and protective effectiveness against lethal A(H7N9) infection in mice. Antibodies that inhibit NA are associated with resistance to influenza virus (10, 13) and correlate with protection against pandemic influenza viruses (15, 33). NA-specific antibodies against N1 and N2 subtypes protect against lethal influenza virus challenge in mice and ferrets (23, 34–38). Consistent with what is known about NA function (12, 39) and previous findings with N1 MAbs (23), the present study shows that N9 MAbs do not interfere with initial virus entry but inhibit virus release from infected cells. SEM observation provided direct evidence that N9 MAbs inhibited virus spread by blocking the release of virions from infected cells. While we cannot completely rule out a contribution by Fc receptor (FcR)-mediated immune effectors, given the poor protection afforded by MAb 11B2, which is a weak inhibitor of NA activity, inhibition of virus spread is likely the primary mechanism that contributed to reduced virus load and protection of N9 MAb-treated mice from death.
In addition to the enzyme active center, there is an HB site on certain subtypes of NA which can bind sialic acids. The HB site on the novel A(H7N9) virus NA was recently reported to enhance binding to human-like sialic acid receptors (26). Our results suggest that this HB site is an ideal target for N9-specific antibodies, with 4 of the 10 MAbs we characterized selecting mutations at residues 434 and/or 371 that are within this region. Similarly, the majority of G70C N9 MAbs prepared by Webster et al. (17) and the single MAb against the novel A(H7N9) generated by Wilson et al. (21) also bind to residues within the HB site. It is interesting that MAbs which bind to the HB region, e.g., 1E8 and 10F4, did not block virus infection, implying that the HB site is not required for virus entry. Our data and the findings of Wilson et al. (21) indicate that MAbs to the HB site can protect mice from lethal A(H7N9) virus challenge, and for this reason the HB site may be an attractive target for antibodies selected for development as antivirals.
As has been shown for G70C NA-specific MAbs (17), antibodies raised against the NA of AH/13 bind to regions that encircle the enzyme active site. The 10 MAbs that we used to select escape mutant viruses identified two major antigenic domains on AH/13 NA. Five of these MAbs (2E6, 2F6, 5C9, 7A4, and 11B2) revealed N9 residues 247, 250, 251, and 253 to be key contacts for antibody binding. These residues are on the outer edge of the NA head and are in an antigenic region also identified in other NA subtypes. In a previous investigation of the antigenic structure of subtype N1, we identified a group of antibodies recognizing the NA of seasonal H1N1 virus A/Brisbane/59/2007, which selected mutations at residues 248, 249, and 250 (23). Also, antibody CD6, which is specific for the NA of the 2009 pandemic H1N1 virus, contacts the 250-loop region through its light chain (34). Another major antigenic domain involves residues in the 370, 400, and 430 loops. Escape mutant selection and site mutagenesis in our study identified residues 368, 370, 371, 373, 400, and 434 as critical contacts for MAbs 1E8, 5H11, 7F12, and 10F4. These amino acids are also within defined epitopes recognized by G70C NA-specific MAbs (17, 20, 21, 40). Amino acid 345 appears to be relatively distant from other identified amino acids but was selected by MAb 5H11, which also selected the K434E escape mutant, suggesting that the footprint of 5H11 covers this entire area. As the majority of the 10 tested MAbs bind to the 250-loop and 370/400/430-loop antigenic domains, it is likely that these two regions are dominant in eliciting an immune response in mice; however, whether they are also the focus of the human antibody response remains to be investigated.
Our results bring to light the unique fine specificity of MAbs that bind to a similar antigenic domain. MAbs 1E8, 5H11, 7F12, and 10F4 all selected escape variants with a mutation at NA residue 434; however, they react differently with NAs containing other mutations (e.g., those at residues 368, 370, 371, 373, and 400) in this region and differ in their in vitro functional attributes. Similarly, MAbs that target the 250-loop domain exhibit different in vitro properties. For example, 11B2 is a poor inhibitor of enzyme activity in ELLAs compared to other MAbs that bind this antigenic domain and was less effective than 2F6 in reducing virus spread in a plaque assay. The difference in inhibition correlates with antibody avidity; however, fine specificity and differences in steric hindrance imposed by antibodies may also contribute to the impact of these antibodies on NA activity. The variation in fine specificity as well as binding avidity might also contribute to differences in the in vivo effectiveness of the MAbs. Of the MAbs tested in vivo, 10F4, which was the most effective, had the greatest avidity for NA and inhibited cleavage of both large and small substrates. A careful, comprehensive evaluation of the properties of MAbs that includes animal challenge studies is warranted before MAbs are selected for further development as prophylactic and therapeutic agents.
Our study highlights the value of NA-specific antibodies in inhibiting replication of the novel A(H7N9) virus, especially in light of antigenic changes that have occurred in the HA of A(H7N9) viruses (41). Both HA and NA of influenza viruses experience antigenic drift; however, there is discordance in their evolution (42), and NA tends to evolve at a lower rate (43, 44). Consistent with this concept, the majority of MAbs characterized in this study cross-react with the NA of an H11N9 virus (G70C) isolated ∼30 years ago although the AH/13 and G70C NAs have as many as 32 amino acid differences in addition to a deletion of amino acids 69 to 73 in AH/13 NA (1). Our sequence analysis revealed that the majority (8 of 12) of the critical residues recognized by our MAbs are conserved in AH/13 and A(H7N9) viruses isolated during the fifth epidemic wave, which occurred between 2016 and 2017. This explains why most of our MAbs also recognized the NA of the most recent A(H7N9) viruses, e.g., GD/16. A major concern raised during treatment of A(H7N9) virus infection has been the emergence of viruses that are resistant to antivirals, including the NA inhibitor oseltamivir. This has indeed occurred; however, the mutations in NA that confer drug resistance did not reduce its sensitivity to N9 MAbs in plaque assays, suggesting that N9 MAbs may provide effective prophylactic and potentially therapeutic treatments should drug-resistant mutations become predominant in the A(H7N9) virus population.
The findings from our study imply the value of N9 antibodies in protection against A(H7N9) infection. Recent studies have demonstrated that live attenuated A(H7N9) vaccine, inactivated cell-cultured whole-virus A(H7N9) vaccine, and virus-like particles bearing HA, NA, and M1 of A(H7N9) virus all induce significant N9 antibody titers in animal models (45–47). The contribution of NA immunity in ameliorating human A(H7N9) infections deserves further investigation in order to improve A(H7N9) vaccines.
In summary, we have prepared and characterized a panel of MAbs recognizing the NA of A(H7N9) viruses that have killed hundreds of individuals in recent years. These antibodies bind to different epitopes within two major antigenic domains on N9 and differ in their abilities to inhibit NA enzyme activity and virus spread. MAb 10F4 that binds to NA with high avidity and inhibits cleavage of both large and small substrates was the most effective in protecting mice against lethal A(H7N9) challenge. Our study highlights differences in the properties of N9-specific MAbs and demonstrates the potential of using NA-specific MAbs in prophylaxis against A(H7N9) virus infection. Further development of these MAbs for use in clinical studies is warranted.
MATERIALS AND METHODS
Viruses.
Reassortant H6N9AH/13 and H6N9GD/16 viruses were generated as previously reported (48, 49). Briefly, pHW2000 plasmids (1 μg each) containing each of the eight virus genes were transfected into a mixture of 293T cells and MDCK cells. The transfection mixture was replaced with Opti-MEM I medium (Invitrogen, Grand Island, NY) after 6 h of incubation at 37°C. Opti-MEM I medium supplemented with trypsin (1 μg/ml) (Sigma-Aldrich, St. Louis, MO) was added 30 h later. The culture supernatant was collected at 48 to 72 h after addition of the transfection mixture, and virus was propagated in 11-day-old embryonated specific-pathogen-free chicken eggs. The wt AH/13 virus was grown in chicken eggs in enhanced biosafety level 3 (BSL3+/ABSL3+) facilities at NIAID, NIH.
Site-directed mutagenesis.
Nucleotide changes corresponding to antiviral-resistant mutations were introduced into the AH/13 NA gene in plasmid pHW2000, and single amino acid mutations in the HB site were introduced into the AH/13 NA gene in the pCAGGS-AH/13 NA plasmid with a QuikChange multisite-directed mutagenesis kit (Stratagene, La Jolla, CA). The mutant plasmids were sequenced to verify the presence of the expected mutations. Reassortant H6N9AH/13 viruses carrying these mutations were rescued by reverse genetics.
MAbs.
To generate mouse hybridomas that secrete N9-specific MAbs, BALB/c mice were immunized and boosted with recombinant AH/13 NA (Sino Biological, Beijing, China). The fusion of splenocytes with mouse myeloma cells Sp2/0 and screening of N9-specific MAbs were performed as previously described (23). Selected hybridomas were cultured in a CELLine device (BD Biosciences, San Jose, CA), and MAbs were purified using protein G columns (GE Healthcare, Uppsala, Sweden). The reactivity of MAbs with N9 was confirmed by ELISA, using NA that was transiently expressed on 293T cells by transfecting cells with pCAGGS plasmid containing the NA gene of AH/13 or G70C (23, 34, 50) or purified H6N9GD/16 virus. The residues on N9 that are critical for MAb binding were identified by selecting antibody escape mutants of H6N9AH/13 virus and sequencing the NA gene of the resultant viruses (as described in the following paragraph) or by performing ELISAs with plasmid-transfected 293T cells (23) or purified parent H6N9AH/13 virus and escape mutants. The values of the optical density at 490 nm (OD490) were normalized to those obtained with wt AH/13 N9 transiently expressed on 293T cells or with the parent H6N9AH/13 virus and expressed as relative binding.
Escape mutants.
The selection of escape mutants with N9 MAbs was conducted according to previously described procedures (23, 34). Briefly, 0.5 ml of each MAb (≥2 mg/ml) was mixed with ∼106 PFU of H6N9AH/13 virus, incubated at room temperature for 30 min, and inoculated into 11-day-old embryonated specific-pathogen-free chicken eggs (5 eggs/MAb). H6N9AH/13 was used to generate escape mutants to keep the consistency of using H6 reassortant viruses in ELLAs (51). After 3 days of incubation, allantoic fluid was collected, and if positive for hemagglutination, it was inoculated onto MDCK cells for plaque assay, with the selecting MAb being supplemented in the agar overlay (1 to 3 μg/ml). Plaques larger than those formed by the parent H6N9AH/13 virus in the presence of the selecting MAb were picked and expanded in eggs. The initial selection with MAbs 1E8, 7F8, and 11B2 did not result in escape mutants, and therefore a second round of selection was performed. The NA gene of each expanded mutant virus was amplified by reverse transcription-PCR (RT-PCR), and PCR products were sequenced. The selection was performed in the same manner with the control antibody 3A2 to confirm that mutations detected in the presence of N9-specific MAbs were not the result of passaging the virus.
NI assays.
The inhibition of NA activity by N9 MAbs was measured by ELLA and MU-NANA assay (51, 52). In the MU-NANA assay, mixtures of a predetermined amount of H6N9AH/13 virus and serial dilutions of antibody were incubated in a 96-well plate. MU-NANA (Sigma-Aldrich, St. Louis, MO) was added and incubated for 1 h at 37°C. The reaction was stopped by addition of 1 N H2SO4, and the plate was read using an excitation wavelength of 355 nm and an emission wavelength of 460 nm. To conduct ELLAs, viruses were titrated to identify the dilution to use in the assay. Antibodies were serially diluted prior to being transferred to fetuin-coated 96-well plates. The selected dilution of H6N9AH/13 virus was added, and the plate was incubated for 18 to 20 h at 37°C. After the plate was washed, horseradish peroxidase (HRP)-conjugated peanut agglutinin (Sigma-Aldrich, St. Louis, MO) was added, and the plate was incubated at room temperature for 2 h. Substrate (o-phenylenediamine dihydrochloride [OPD]) was added after the plate was washed. The reaction was stopped after a 10-min incubation. OD490 values were measured, and the percent inhibition was calculated after background values were subtracted. In both assays, the inverse of the dilution that resulted in 50% inhibition was calculated by nonlinear regression analysis (GraphPad Prism, version 5.0c; GraphPad Software, Inc., San Diego, CA) and recorded as NI titer.
Plaque assay.
MDCK cells growing in 6- or 12-well plates were infected with diluted virus for 1 h at 37°C. The virus inoculum was removed, and the cells were washed with phosphate-buffered saline (PBS) and overlaid with agar supplemented with MAbs at various concentrations and 1 μg/ml of trypsin. To examine the impact of N9 MAbs on virus binding/entry, virus was premixed with antibodies and incubated at room temperature for 30 min before cells were inoculated. After 1 h of incubation, cells were washed thoroughly with PBS to remove antibodies and covered with agar containing 1 μg/ml of trypsin but without antibody. The cells were incubated at 33°C or 37°C in 5% CO2 for 3 days and then fixed with methanol, followed by staining with crystal violet solution to visualize the plaques. MAb 3A2 (23) was used as a negative control. To compare the sizes (diameters) of the plaques, ≥10 plaques were randomly selected, the plaque images were magnified in Photoshop software (CS6 extended), and the sizes were determined using the measurement tool within the software and then converted to the real size (in millimeters).
SEM observation.
MDCK cells growing on coverslips (Neuvitro, Vancouver, WA) in 12-well plates were infected with H6N9AH/13 virus at a multiplicity of infection (MOI) of 3, followed by incubation with medium supplemented with N9 MAbs or the control antibody 3A2 at 10 μg/ml. Cells were fixed with 2.5% glutaraldehyde and processed for observation with SEM. Briefly, the fixed cells were washed with 0.1 M sodium cacodylate buffer (pH 7.4), followed by postfixation with 1% OsO4 in PBS at room temperature for 1 h. After cells were washed with water, they were dehydrated in 50%, 70%, 95%, and absolute ethanol and then transferred to a pressure chamber-critical point dryer (Leica EM CPD 300), followed by coating with 12-nm gold in high-resolution sputter (Leica ACE 200). The processed samples were examined with Tescan Mira3 FE-SEM, and images were acquired using a Tescan digital camera.
Affinity measurement by SPR.
Steady-state equilibrium binding of serial dilutions of MAbs was monitored at 25°C using a ProteOn SPR biosensor (Bio-Rad, Hercules, CA). Five dilutions of AH/13 NA (Sino Biological, Beijing, China) were coupled to a GLC sensor chip with amine coupling in the test flow cells. Samples of freshly prepared antibody (200 μl) at 5-fold dilutions (starting concentrations of 50 μg/ml for MAbs 1E8, 5H11, and 7F12 and of 10 μg/ml for other MAbs) were injected at a flow rate of 50 μl/min (120-s contact time) for association, and dissociation was performed over a 600-s interval (at a flow rate of 50 μl/min). Responses from the protein surface were corrected for the response from a mock surface and for measurements from a separate, buffer-only injection as described before (27, 28). Binding kinetics for the antibody-antigen interaction was calculated by a bivalent analyte model using Bio-Rad ProteOn manager software (version 3.0.1). To improve the measurements, the KDs were determined from two independent SPR runs.
Animal study.
Female BALB/c mice (8 weeks old; The Jackson Laboratory, Bar Harbor, ME) were used in the study, which followed federal guidelines and protocols approved by the Institutional Animal Care and Use Committees (CBER, FDA and NIAID, NIH). To determine the prophylactic efficacy, groups of mice (n = 10) were treated with antibodies 2F6, 10F4, and 11B2 at doses of 0.5 and 5 mg/kg or with the control antibody 3A2 at 5 mg/kg. All doses were administered i.p. in a volume of 200 μl in PBS. Approximately 12 h later, the mice were challenged i.n. with 10 MLD50 of AH/13 in 50 μl of PBS. On day 6 p.c., five mice from each group were euthanized. The lungs from three mice were collected for virus titration in MDCK cells, and the lungs from two mice were fixed with 10% formalin, embedded in paraffin, sectioned, and stained with H&E. The remaining mice (5 per group) were monitored daily for up to 14 days for survival and weight loss. Mice that lost ≥25% of weight were euthanized. All experiments with live, wt A(H7N9) virus (animal challenge and monitoring, sample collection, virus titration, etc.) were conducted in enhanced biosafety level 3 (BSL3+/ABSL3+) facilities at NIAID, NIH. The tissues for histology were fixed and processed according to approved protocols.
Sequence alignment.
To examine the variation in sequence at the antigenically critical amino acid positions, 169 full-length NA sequences of human A(H7N9) viruses isolated between 1 October 2016 and 20 October 2017 were obtained from GISAID (http://www.gisaid.org) and compared to the AH/13 NA sequence using the positional frequency summary feature of BioEdit software (version 7.2.5 [http://www.mbio.ncsu.edu/BioEdit]). The variation was expressed as the percentage of viruses carrying a different residue at each critical amino acid position identified in this study.
Statistical analysis.
Data were analyzed by using GraphPad Prism, version 5.0c (GraphPad Software, Inc., San Diego, CA). One-way or two-way analysis of variance (ANOVA) was applied, and Tukey multiple comparison was used for post hoc analysis. Probability values of ≤0.05 were used to indicate a statistically significant difference.
ACKNOWLEDGMENTS
This study was supported by the FDA and by the intramural research program of the NIAID, NIH.
We thank Robert G. Webster (St. Jude Children's Research Hospital, Memphis, TN) and Zhiping Ye and Carol Weiss (CBER, FDA) for providing plasmids used in generating H6N9 viruses and cell-based ELISA, Larisa Gubareva, Vasiliy Mishin (Influenza Division, CDC), and Xing Li (CBER, FDA) for helpful technical discussions, and Jianqiang Ye (College of Veterinary Medicine, Yangzhou University, China) for assistance in analyzing the NA sequences. We also thank Yong Wu and Jiwen Zheng (CDRH, FDA) for their permission to use the SEM. We are indebted to the veterinary service staff at CBER, FDA, and NIAID, NIH, for excellent animal care and the Facility for Biotechnology Resources, CBER, FDA, for DNA sequencing.
REFERENCES
- 1.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 2.Jernigan DB, Cox NJ. 2015. H7N9: preparing for the unexpected in influenza. Annu Rev Med 66:361–371. doi: 10.1146/annurev-med-010714-112311. [DOI] [PubMed] [Google Scholar]
- 3.Liu D, Zhang Z, He L, Gao Z, Li J, Gu M, Hu J, Wang X, Liu X, Liu X. 21 September 2017. Characteristics of the emerging chicken-origin highly pathogenic H7N9 viruses: a new threat to public health and poultry industry. J Infect doi: 10.1016/j.jinf.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Madan A, Segall N, Ferguson M, Frenette L, Kroll R, Friel D, Soni J, Li P, Innis BL, Schuind A. 2016. Immunogenicity and safety of an AS03-adjuvanted H7N9 pandemic influenza vaccine in a randomized trial in healthy adults. J Infect Dis 214:1717–1727. doi: 10.1093/infdis/jiw414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulligan MJ, Bernstein DI, Winokur P, Rupp R, Anderson E, Rouphael N, Dickey M, Stapleton JT, Edupuganti S, Spearman P, Ince D, Noah DL, Hill H, Bellamy AR, DMID 13-0032 H7N9 Vaccine Study Group . 2014. Serological responses to an avian influenza A/H7N9 vaccine mixed at the point-of-use with MF59 adjuvant: a randomized clinical trial. JAMA 312:1409–1419. doi: 10.1001/jama.2014.12854. [DOI] [PubMed] [Google Scholar]
- 6.Sobhanie M, Matsuoka Y, Jegaskanda S, Fitzgerald T, Mallory R, Chen Z, Luke C, Treanor J, Subbarao K. 2016. Evaluation of the safety and immunogenicity of a candidate pandemic live attenuated influenza vaccine (pLAIV) against influenza A(H7N9). J Infect Dis 213:922–929. doi: 10.1093/infdis/jiv526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen M, Zhang XQ, Senaati HP, Chen HW, Varki NM, Schooley RT, Gagneux P. 2013. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J 10:321. doi: 10.1186/1743-422X-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu W, Zhou J, Li Z, Yang L, Li X, Huang W, Zou S, Chen W, Wei H, Tang J, Liu L, Dong J, Wang D, Shu Y. 2017. Biological characterisation of the emerged highly pathogenic avian influenza (HPAI) A(H7N9) viruses in humans, in mainland China, 2016 to 2017. Euro Surveill 22:30533. doi: 10.2807/1560-7917.ES.2017.22.19.30533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shtyrya YA, Mochalova LV, Bovin NV. 2009. Influenza virus neuraminidase: structure and function. Acta Naturae 1:26–32. [PMC free article] [PubMed] [Google Scholar]
- 10.Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, Davey RT Jr, Taubenberger JK. 2016. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. mBio 7:e00417-16. doi: 10.1128/mBio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cate TR, Rayford Y, Nino D, Winokur P, Brady R, Belshe R, Chen W, Atmar RL, Couch RB. 2010. A high dosage influenza vaccine induced significantly more neuraminidase antibody than standard vaccine among elderly subjects. Vaccine 28:2076–2079. doi: 10.1016/j.vaccine.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichelberger MC, Wan H. 2015. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol 386:275–299. [DOI] [PubMed] [Google Scholar]
- 13.Murphy BR, Kasel JA, Chanock RM. 1972. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N Engl J Med 286:1329–1332. doi: 10.1056/NEJM197206222862502. [DOI] [PubMed] [Google Scholar]
- 14.Couch RB, Kasel JA, Gerin JL, Schulman JL, Kilbourne ED. 1974. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. J Infect Dis 129:411–420. doi: 10.1093/infdis/129.4.411. [DOI] [PubMed] [Google Scholar]
- 15.Monto AS, Kendal AP. 1973. Effect of neuraminidase antibody on Hong Kong influenza. Lancet i:623–625. [DOI] [PubMed] [Google Scholar]
- 16.Beutner KR, Chow T, Rubi E, Strussenberg J, Clement J, Ogra PL. 1979. Evaluation of a neuraminidase-specific influenza A virus vaccine in children: antibody responses and effects on two successive outbreaks of natural infection. J Infect Dis 140:844–850. doi: 10.1093/infdis/140.6.844. [DOI] [PubMed] [Google Scholar]
- 17.Webster RG, Air GM, Metzger DW, Colman PM, Varghese JN, Baker AT, Laver WG. 1987. Antigenic structure and variation in an influenza virus N9 neuraminidase. J Virol 61:2910–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Air GM, Webster RG, Colman PM, Laver WG. 1987. Distribution of sequence differences in influenza N9 neuraminidase of tern and whale viruses and crystallization of the whale neuraminidase complexed with antibodies. Virology 160:346–354. doi: 10.1016/0042-6822(87)90005-5. [DOI] [PubMed] [Google Scholar]
- 19.Tulip WR, Harley VR, Webster RG, Novotny J. 1994. N9 neuraminidase complexes with antibodies NC41 and NC10: empirical free energy calculations capture specificity trends observed with mutant binding data. Biochemistry 33:7986–7997. doi: 10.1021/bi00192a002. [DOI] [PubMed] [Google Scholar]
- 20.Colman PM, Laver WG, Varghese JN, Baker AT, Tulloch PA, Air GM, Webster RG. 1987. Three-dimensional structure of a complex of antibody with influenza virus neuraminidase. Nature 326:358–363. doi: 10.1038/326358a0. [DOI] [PubMed] [Google Scholar]
- 21.Wilson JR, Guo Z, Reber A, Kamal RP, Music N, Gansebom S, Bai Y, Levine M, Carney P, Tzeng WP, Stevens J, York IA. 2016. An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody with prophylactic and therapeutic activity in vivo. Antiviral Res 135:48–55. doi: 10.1016/j.antiviral.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohler G, Milstein C. 1976. Derivation of specific antibody-producing tissue culture and tumor lines by cell fusion. Eur J Immunol 6:511–519. doi: 10.1002/eji.1830060713. [DOI] [PubMed] [Google Scholar]
- 23.Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, Ye J, Sultana I, Wan XF, Liu X, Perez DR, Taubenberger JK, Eichelberger MC. 2013. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol 87:9290–9300. doi: 10.1128/JVI.01203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nuss JM, Air GM. 1991. Transfer of the hemagglutinin activity of influenza virus neuraminidase subtype N9 into an N2 neuraminidase background. Virology 183:496–504. doi: 10.1016/0042-6822(91)90979-L. [DOI] [PubMed] [Google Scholar]
- 25.Laver WG, Colman PM, Webster RG, Hinshaw VS, Air GM. 1984. Influenza virus neuraminidase with hemagglutinin activity. Virology 137:314–323. doi: 10.1016/0042-6822(84)90223-X. [DOI] [PubMed] [Google Scholar]
- 26.Benton DJ, Wharton SA, Martin SR, McCauley JW. 2017. Role of neuraminidase in influenza A(H7N9) virus receptor binding. J Virol 91:e02293-16. doi: 10.1128/JVI.02293-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khurana S, Verma N, Yewdell JW, Hilbert AK, Castellino F, Lattanzi M, Del Giudice G, Rappuoli R, Golding H. 2011. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med 3:85ra48. doi: 10.1126/scitranslmed.3002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khurana S, Fuentes S, Coyle EM, Ravichandran S, Davey RT Jr, Beigel JH. 2016. Human antibody repertoire after VSV-Ebola vaccination identifies novel targets and virus-neutralizing IgM antibodies. Nat Med 22:1439–1447. doi: 10.1038/nm.4201. [DOI] [PubMed] [Google Scholar]
- 29.Marjuki H, Mishin VP, Chesnokov AP, De La Cruz JA, Davis CT, Villanueva JM, Fry AM, Gubareva LV. 2015. Neuraminidase mutations conferring resistance to oseltamivir in influenza A(H7N9) viruses. J Virol 89:5419–5426. doi: 10.1128/JVI.03513-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, Lu S, Song Z, Wang W, Hao P, Li J, Zhang X, Yen HL, Shi B, Li T, Guan W, Xu L, Liu Y, Wang S, Zhang X, Tian D, Zhu Z, He J, Huang K, Chen H, Zheng L, Li X, Ping J, Kang B, Xi X, Zha L, Li Y, Zhang Z, Peiris M, Yuan Z. 2013. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet 381:2273–2279. doi: 10.1016/S0140-6736(13)61125-3. [DOI] [PubMed] [Google Scholar]
- 31.Marjuki H, Mishin VP, Chesnokov AP, Jones J, De La Cruz JA, Sleeman K, Tamura D, Nguyen HT, Wu HS, Chang FY, Liu MT, Fry AM, Cox NJ, Villanueva JM, Davis CT, Gubareva LV. 2015. Characterization of drug-resistant influenza A(H7N9) variants isolated from an oseltamivir-treated patient in Taiwan. J Infect Dis 211:249–257. doi: 10.1093/infdis/jiu447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang N, Li X, Ren R, Wang D, Zhou S, Greene CM, Song Y, Zhou L, Yang L, Davis CT, Zhang Y, Wang Y, Zhao J, Li X, Iuliano AD, Havers F, Olsen SJ, Uyeki TM, Azziz-Baumgartner E, Trock S, Liu B, Sui H, Huang X, Zhang Y, Ni D, Feng Z, Shu Y, Li Q. 2016. Assessing change in avian influenza A(H7N9) virus infections during the fourth epidemic—China, September 2015–August 2016. MMWR Morb Mortal Wkly Rep 65:1390–1394. doi: 10.15585/mmwr.mm6549a2. [DOI] [PubMed] [Google Scholar]
- 33.Marcelin G, DuBois R, Rubrum A, Russell CJ, McElhaney JE, Webby RJ. 2011. A contributing role for anti-neuraminidase antibodies on immunity to pandemic H1N1 2009 influenza A virus. PLoS One 6:e26335. doi: 10.1371/journal.pone.0026335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan H, Yang H, Shore DA, Garten RJ, Couzens L, Gao J, Jiang L, Carney PJ, Villanueva J, Stevens J, Eichelberger MC. 2015. Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers. Nat Commun 6:6114. doi: 10.1038/ncomms7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyle TM, Hashem AM, Li C, Van Domselaar G, Larocque L, Wang J, Smith D, Cyr T, Farnsworth A, He R, Hurt AC, Brown EG, Li X. 2013. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antiviral Res 100:567–574. doi: 10.1016/j.antiviral.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 36.Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, Brokstad KA, Cox RJ, Palese P, Krammer F. 2015. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. mBio 6:e02556-14. doi: 10.1128/mBio.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monto AS, Petrie JG, Cross RT, Johnson E, Liu M, Zhong W, Levine M, Katz JM, Ohmit SE. 2015. Antibody to influenza virus neuraminidase: an independent correlate of protection. J Infect Dis 212:1191–1199. doi: 10.1093/infdis/jiv195. [DOI] [PubMed] [Google Scholar]
- 38.Cardoso FM, Ibanez LI, Van den Hoecke S, De Baets S, Smet A, Roose K, Schepens B, Descamps FJ, Fiers W, Muyldermans S, Depicker A, Saelens X. 2014. Single-domain antibodies targeting neuraminidase protect against an H5N1 influenza virus challenge. J Virol 88:8278–8296. doi: 10.1128/JVI.03178-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colman PM. 1994. Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein Sci 3:1687–1696. doi: 10.1002/pro.5560031007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colman PM, Tulip WR, Varghese JN, Tulloch PA, Baker AT, Laver WG, Air GM, Webster RG. 1989. Three-dimensional structures of influenza virus neuraminidase-antibody complexes. Philos Trans R Soc Lond B Biol Sci 323:511–518. doi: 10.1098/rstb.1989.0028. [DOI] [PubMed] [Google Scholar]
- 41.Iuliano AD, Jang Y, Jones J, Davis CT, Wentworth DE, Uyeki TM, Roguski K, Thompson MG, Gubareva L, Fry AM, Burns E, Trock S, Zhou S, Katz JM, Jernigan DB. 2017. Increase in human infections with avian influenza A(H7N9) virus during the fifth epidemic—China, October 2016–February 2017. MMWR Morb Mortal Wkly Rep 66:254–255. doi: 10.15585/mmwr.mm6609e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, Burke DF, Smith DJ, Fouchier RA, Eichelberger MC. 2011. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc Natl Acad Sci U S A 108:20748–20753. doi: 10.1073/pnas.1113801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kilbourne ED, Johansson BE, Grajower B. 1990. Independent and disparate evolution in nature of influenza A virus hemagglutinin and neuraminidase glycoproteins. Proc Natl Acad Sci U S A 87:786–790. doi: 10.1073/pnas.87.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein EY, Serohijos AW, Choi JM, Shakhnovich EI, Pekosz A. 2014. Influenza A H1N1 pandemic strain evolution—divergence and the potential for antigenic drift variants. PLoS One 9:e93632. doi: 10.1371/journal.pone.0093632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Jonge J, Isakova-Sivak I, van Dijken H, Spijkers S, Mouthaan J, de Jong R, Smolonogina T, Roholl P, Rudenko L. 2016. H7N9 live attenuated influenza vaccine is highly immunogenic, prevents virus replication, and protects against severe bronchopneumonia in ferrets. Mol Ther 24:991–1002. doi: 10.1038/mt.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wodal W, Schwendinger MG, Savidis-Dacho H, Crowe BA, Hohenadl C, Fritz R, Bruhl P, Portsmouth D, Karner-Pichl A, Balta D, Grillberger L, Kistner O, Barrett PN, Howard MK. 2015. Immunogenicity and protective efficacy of a Vero cell culture-derived whole-virus H7N9 vaccine in mice and guinea pigs. PLoS One 10:e0113963. doi: 10.1371/journal.pone.0113963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu CJ, Chien CY, Liu MT, Fang ZS, Chang SY, Juang RH, Chang SC, Chen HW. 2017. Multi-antigen avian influenza a (H7N9) virus-like particles: particulate characterizations and immunogenicity evaluation in murine and avian models. BMC Biotechnol 17:2. doi: 10.1186/s12896-016-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan H, Perez DR. 2007. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J Virol 81:5181–5191. doi: 10.1128/JVI.02827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A 96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Ilyushina NA, Lugovtsev VY, Bovin NV, Couzens LK, Gao J, Donnelly RP, Eichelberger MC, Wan H. 2017. Amino acids in hemagglutinin antigenic site B determine antigenic and receptor binding differences between A(H3N2)v and ancestral seasonal H3N2 influenza viruses. J Virol 91:e01512-16. doi: 10.1128/JVI.01512-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Couzens L, Gao J, Westgeest K, Sandbulte M, Lugovtsev V, Fouchier R, Eichelberger M. 2014. An optimized enzyme-linked lectin assay to measure influenza A virus neuraminidase inhibition antibody titers in human sera. J Virol Methods 210:7–14. doi: 10.1016/j.jviromet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Wan Z, Ye J, Sang J, Shao H, Qian K, Jin W, Qin A, Wan H. 2016. Identification of amino acids in H9N2 influenza virus neuraminidase that are critical for the binding of two mouse monoclonal antibodies. Vet Microbiol 187:58–63. doi: 10.1016/j.vetmic.2016.03.011. [DOI] [PubMed] [Google Scholar]