ABSTRACT
Ferrets and mice are frequently used as animal models for influenza research. However, ferrets are demanding in terms of housing space and handling, whereas mice are not naturally susceptible to infection with human influenza A or B viruses. Therefore, prior adaptation of human viruses is required for their use in mice. In addition, there are no mouse-adapted variants of the recent H3N2 viruses, because these viruses do not replicate well in mice. In this study, we investigated the susceptibility of Syrian hamsters to influenza viruses with a view to using the hamster model as an alternative to the mouse model. We found that hamsters are sensitive to influenza viruses, including the recent H3N2 viruses, without adaptation. Although the hamsters did not show weight loss or clinical signs of H3N2 virus infection, we observed pathogenic effects in the respiratory tracts of the infected animals. All of the H3N2 viruses tested replicated in the respiratory organs of the hamsters, and some of them were detected in the nasal washes of infected animals. Moreover, a 2009 pandemic (pdm09) virus and a seasonal H1N1 virus, as well as one of the two H3N2 viruses, but not a type B virus, were transmissible by the airborne route in these hamsters. Hamsters thus have the potential to be a small-animal model for the study of influenza virus infection, including studies of the pathogenicity of H3N2 viruses and other strains, as well as for use in H1N1 virus transmission studies.
IMPORTANCE We found that Syrian hamsters are susceptible to human influenza viruses, including the recent H3N2 viruses, without adaptation. We also found that a pdm09 virus and a seasonal H1N1 virus, as well as one of the H3N2 viruses, but not a type B virus tested, are transmitted by the airborne route in these hamsters. Syrian hamsters thus have the potential to be used as a small-animal model for the study of human influenza viruses.
KEYWORDS: animal model, hamster, influenza
INTRODUCTION
Influenza A viruses are known to have a broad host range. They can infect not only humans but also waterfowl, poultry, sea mammals, pigs, horses, cats, dogs, and other species (1). Ferrets are used as an experimental animal model for studies of influenza virus infection because they are naturally susceptible to influenza A and B viruses, and their clinical features and the pathological changes associated with the bronchitis and pneumonia that they experience resemble those that occur in humans (2–5). Ferrets have also been used for studies of influenza virus transmission (6–8). However, ferrets demand considerable housing space and can be difficult to handle. Mice are frequently used as an animal model for influenza research. However, mice are not naturally susceptible to human influenza A or B viruses, with the exception of highly pathogenic human H5N1 viruses (9), the reconstructed 1918 pandemic influenza virus (10), the A(H1N1) pandemic 2009 [A(H1N1)pdm09] virus (6, 11), and H7N9 viruses (8, 12–14). Therefore, prior adaptation of human viruses is required for their experimental use in mice. Other rodents, such as rats, guinea pigs, and cotton rats, are also occasionally used as animal models. To use rats experimentally, rat-adapted viruses, which induce a mild form of the disease with no mortality, are required (15). Guinea pigs have been used as a model of transmission of influenza viruses (16); however, even though the H5N1 and 1918 pandemic viruses replicated in the lungs and nasal turbinates of these animals, no weight loss or morbidity was observed (17). Recently, cotton rats have been considered a potential animal model for influenza viruses (18). They are susceptible to both human influenza A and B viruses without prior adaptation (19–22), and an H5N1 virus was shown to be lethal in this species (22). However, cotton rats are not widely available. Thus, each animal model has limitations or drawbacks.
Current animal models for influenza virus studies have one additional limitation. Historically, H3N2 viruses, such as A/Hong Kong/1/68, A/Aichi/2/68, and A/Guizhou/54/89, were adapted to mice, and the mouse-adapted variants were used for numerous studies (23–26). However, the recent H3N2 viruses cannot replicate in mice (27), which prevents mouse models from being used to test the activities of therapeutic or prophylactic drugs against the currently prevalent viruses.
Previously, hamsters were proposed to be a model animal for the study of influenza, because of their sensitivity to human isolates (28–33) and contact transmission of human isolates (34). Hamsters showed sensitivity equivalent to that of ferrets and guinea pigs (33). Moreover, in a vaccine efficacy study, hamsters differentially recognized a single amino acid difference involving egg adaptation in the H1 hemagglutinin (HA) protein (33). In this study, we investigated the susceptibility of Syrian hamsters to influenza viruses to assess the possibility of using these animals as a small-animal model for influenza research.
RESULTS
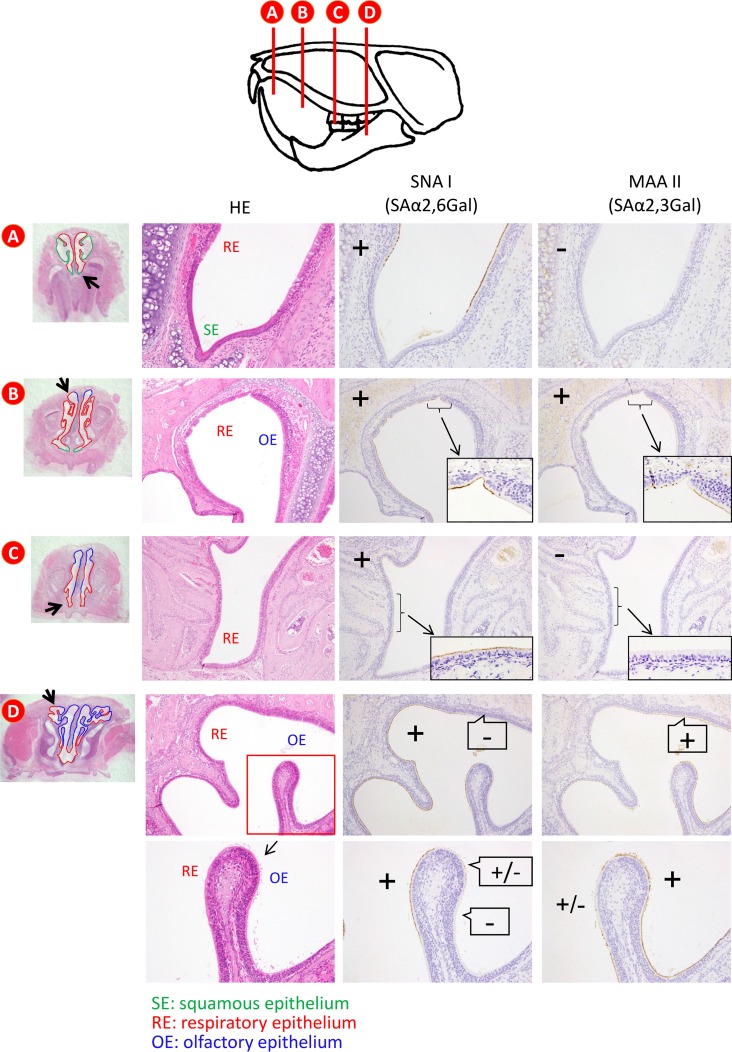
Detection of sialyloligosaccharides in the respiratory tract of hamsters.
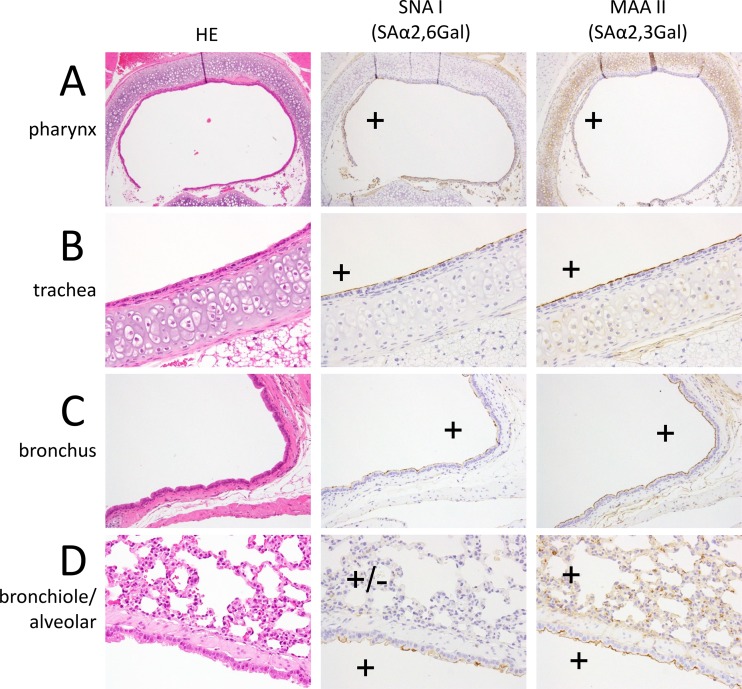
First, we examined the sialyloligosaccharide distribution in the respiratory tract of 4- and 8-week-old female hamsters. The nasal epithelial cell populations varied at different locations (35, 36). At the distal section of the nasal cavity of 4-week-old hamsters, squamous epithelial and respiratory epithelial cells predominated (Fig. 1A). The population of olfactory epithelial cells gradually increased from the middle to the deep section of the nasal cavity (Fig. 1B and C), such that there were ultimately more olfactory epithelial cells than respiratory epithelial cells in the deep portion of the nasal cavity (Fig. 1D). Sambucus nigra lectin I (SNA I), which is specific for sialic acid linked to galactose by an α-2,6 linkage (SAα2,6Gal), mainly reacted with the respiratory epithelial cells in the distal section of the nasal cavity (Fig. 1A to C); in contrast, Maackia amurensis lectin II (MAA II), which is specific for sialic acid linked to galactose by an α-2,3 linkage (SAα2,3Gal), mainly reacted with the olfactory epithelial cells in the proximal portion of the nasal turbinates of the hamsters (Fig. 1C and D). In the pharynx, trachea, and bronchus, both SNA I and MAA II strongly reacted with the epithelial cells (Fig. 2A to C). In contrast, only MAA II strongly reacted with the epithelial cells in the lungs (Fig. 2D). Similar findings were obtained with the 8-week-old hamsters (data not shown). These results indicate that 4- and 8-week-old hamsters have appreciable amounts of SAα2,6Gal in the distal end of their nasal turbinates and SAα2,3Gal in their lungs.
FIG 1.
Detection of SAα2,6Gal and SAα2,3Gal oligosaccharides in the nasal turbinate by using lectins. Sections of a 4-week-old Syrian hamster were reacted with SNA I and MAA II. The vertical lines of the image at the top indicate the anterior surfaces of transverse tissue blocks (A to D). (A) A distal section of the nasal cavity of a 4-week-old hamster showing the predominance of squamous epithelial cells and respiratory epithelial cells. (B to D) The population of olfactory epithelial cells gradually increased from the middle to the deep section of the nasal cavity (B, C); more olfactory epithelial cells than respiratory epithelial cells were present in the deep portion of the nasal cavity (D). SNA I, which is specific for SAα2,6Gal, mainly reacted with respiratory epithelial cells in the distal section of the nasal cavity (A to C); in contrast, MAA II, which is specific for SAα2,3Gal, mainly reacted with olfactory epithelial cells in the proximal portion of the nasal turbinates of hamsters (C, D). HE, hematoxylin and eosin staining.
FIG 2.
Detection of SAα2,6Gal and SAα2,3Gal oligosaccharides in the pharynx (A), trachea (B), bronchus (C), and bronchiole/alveolar region (D) of a 4-week-old Syrian hamster. In the pharynx, trachea, and bronchus, both SNA I and MAA II strongly reacted with the epithelial cells (A, B, C). In contrast, MAA II strongly reacted with the epithelial cells in the lungs (D). HE, hematoxylin and eosin staining.
Growth properties of H3N2 viruses in hamsters and mice.
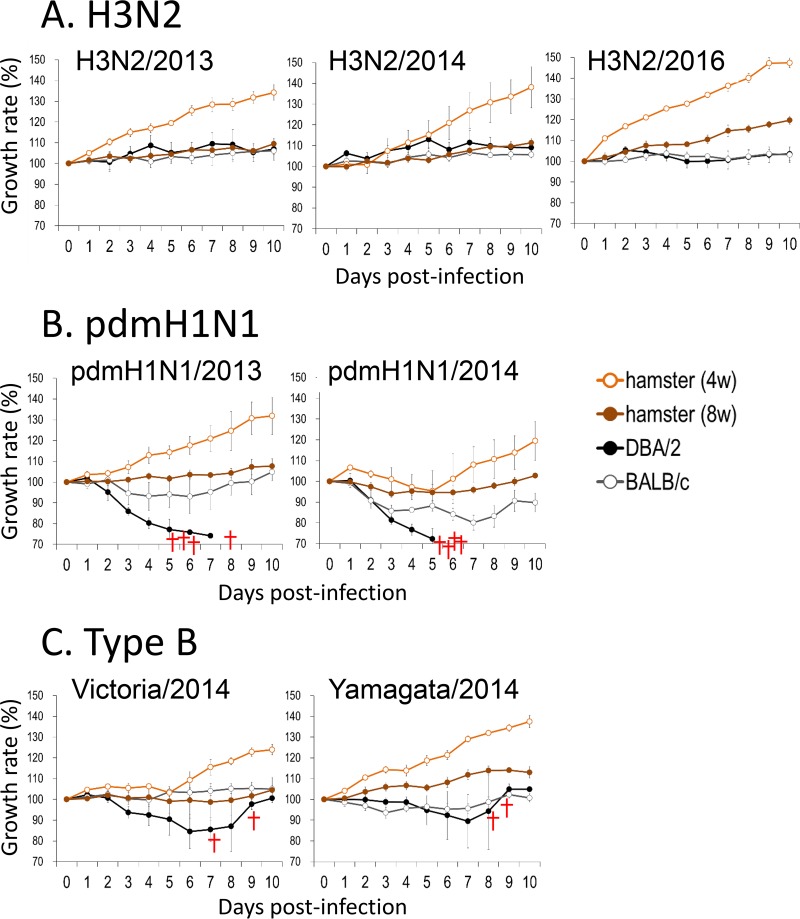
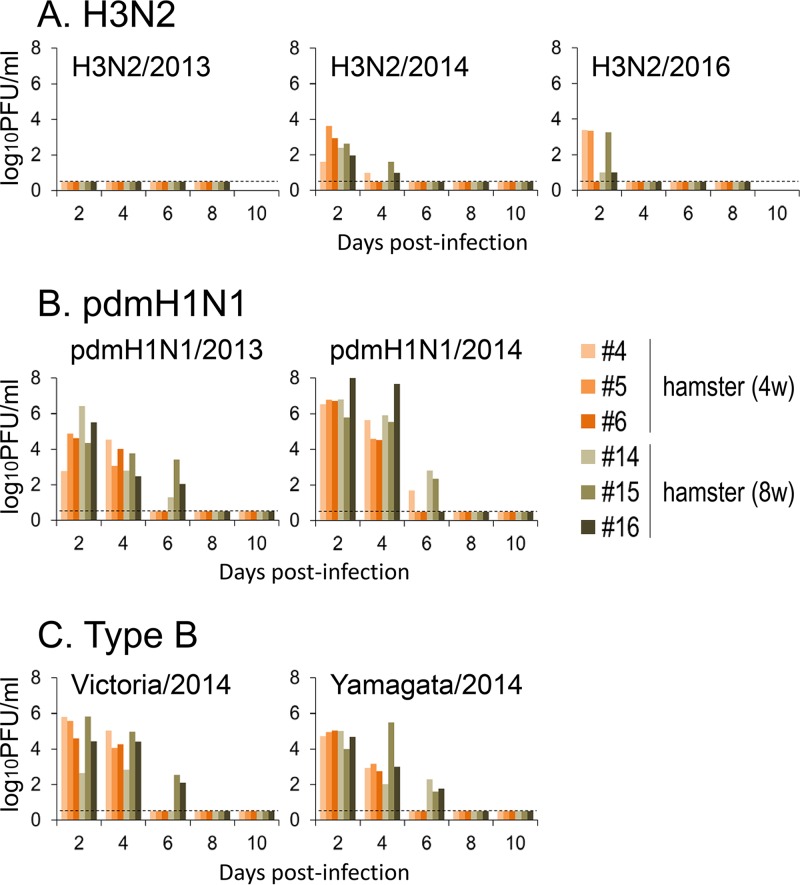
Four- or 8-week-old female hamsters and 6-week-old female BALB/c or DBA/2 mice were anesthetized and intranasally inoculated with 1.0 × 106 PFU of A/Tokyo/IMS6-1/2013 (H3N2/2013), A/Tokyo/IMS2-1/2014 (H3N2/2014), or A/Tokyo/UT-HP002/2016 (H3N2/2016) virus (n = 9 hamsters and n = 13 mice for each virus). The clinical condition and body weight of 3 hamsters and 4 mice infected with each virus were assessed daily, and nasal wash specimens were collected from the hamsters every other day for virus titration. None of the infected animals showed any clinical signs (data not shown) or weight loss, with the exception of a slight decrease in the body weight of the H3N2/2016-infected mice (Fig. 3A). Although the H3N2/2013 virus was not detected in the nasal washes of H3N2/2013-infected hamsters, viruses were detected in the nasal washes until day 4 in all H3N2/2014-infected hamsters and 5 of 6 H3N2/2016-infected hamsters (Fig. 4A). On days 1 (n = 3, mice only), 3 (n = 3), and 6 (n = 3) postinfection, animals were euthanized and their organs were collected for virological and pathological examination. Although no clinical signs were observed, all of the viruses replicated in the respiratory organs of the hamsters (Table 1). In contrast, among the DBA/2 mice, the H3N2/2013 virus was found in the trachea of only one mouse. H3N2/2014 virus titers were moderate in the nasal turbinates on days 1 and 3, and the virus was found in the lung of one DBA/2 mouse and the trachea of another DBA/2 mouse on day 1 (Table 1). The H3N2/2016 virus was detected at low levels in the lungs, tracheas, and nasal turbinates of DBA/2 mice. Virus titers in BALB/c mice were generally lower than those in DBA/2 mice, as previously reported (37). No virus was detected in any DBA/2 or BALB/c mouse on day 6 (Table 1). These results indicate that hamsters are more susceptible to the recent H3N2 viruses than are BALB/c or DBA/2 mice.
FIG 3.
Body weight changes in infected animals. Six-week-old female BALB/c mice and DBA/2 mice and 4- or 8-week-old female Syrian hamsters were anesthetized and intranasally inoculated with 106 PFU of H3N2/2013 (A, left), H3N2/2014 (A, middle), H3N2/2016 (A, right), pdmH1N1/2013 (B, left), pdmH1N1/2014 (B, right), Victoria/2014 (C, left), or Yamagata/2014 (C, right) virus (n = 3 hamsters and n = 4 mice for each virus). The body weights of individual animals inoculated with viruses are depicted as a percentage of the body weight compared with that on day 0. Crosses indicate dead infected animals.
FIG 4.
Virus titers in the nasal washes of 4-week-old or 8-week-old hamsters infected with 106 PFU of H3N2/2013 (A, left), H3N2/2014 (A, middle), H3N2/2016 (A, right), pdmH1N1/2013 (B, left), pdmH1N1/2014 (B, right), Victoria/2014 (C, left), or Yamagata/2014 (C, right) virus (n = 3 hamsters for each virus). Nasal wash specimens with 400 μl of PBS from each hamster were collected every other day for virus titration. Virus titers were determined by using a plaque assay on MDCK cells. The lower limit of detection is indicated by the horizontal dashed lines.
TABLE 1.
Virus titers in tissues of animals infected with H3N2 virusesa
| Animal (age) and organb | Titer (log10 PFU/g) |
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H3N2/2013 |
H3N2/2014 |
H3N2/2016 |
|||||||||||||||||||||||||
| 24 hpi |
Day 3 |
Day 6 |
24 hpi |
Day 3 |
Day 6 |
24 hpi |
Day 3 |
Day 6 |
|||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | |
| DBA/2 mice | |||||||||||||||||||||||||||
| Nasal turb | —c | — | — | — | — | — | — | — | — | 3.4 | 4.9 | 3.6 | 4.0 | 3.8 | 3.6 | — | — | — | 2.1 | — | — | — | — | — | — | — | — |
| Trachea | 2.3 | — | — | — | — | — | — | — | — | — | — | 2.4 | — | — | — | — | — | — | 2.3 | 3.7 | 3.6 | — | — | — | — | — | — |
| Lung | — | — | — | — | — | — | — | — | — | 1.9 | — | — | — | — | — | — | — | — | 3.3 | 3.7 | 1.8 | — | — | — | — | — | — |
| BALB/c mice | |||||||||||||||||||||||||||
| Nasal turb | — | — | — | — | — | — | — | — | — | 3.0 | 3.7 | — | 3.8 | 2.8 | 4.9 | — | — | — | — | — | — | — | — | — | — | — | — |
| Trachea | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 2.6 | — | — | — | — | — | — | — |
| Lung | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 2.9 | — | — | — | — | — | — |
| Hamsters (4 wk) | |||||||||||||||||||||||||||
| Nasal turb | NAd | NA | NA | 4.0 | 4.5 | 5.7 | — | — | 2.6 | NA | NA | NA | 5.7 | 5.3 | 5.9 | 2.8 | — | 2.8 | NA | NA | NA | 6.0 | 5.7 | 5.8 | — | — | — |
| Trachea | NA | NA | NA | 5.6 | 5.0 | 4.6 | — | — | — | NA | NA | NA | 4.0 | 3.0 | 4.5 | — | — | — | NA | NA | NA | 3.7 | 4.5 | 3.4 | — | — | — |
| Lung | |||||||||||||||||||||||||||
| R cra/acce | NA | NA | NA | 5.2 | 3.4 | — | — | — | — | NA | NA | NA | 2.7 | — | — | — | — | — | NA | NA | NA | 4.9 | 4.6 | — | — | — | — |
| R middle | NA | NA | NA | 4.7 | 4.0 | — | — | — | — | NA | NA | NA | 3.4 | — | — | — | — | — | NA | NA | NA | 4.7 | 5.4 | — | — | — | — |
| R caudal | NA | NA | NA | 5.3 | 3.5 | — | — | — | — | NA | NA | NA | 2.9 | 2.5 | — | — | — | — | NA | NA | NA | 5.8 | 6.1 | — | — | — | — |
| L | NA | NA | NA | 5.5 | 4.3 | — | — | — | — | NA | NA | NA | 2.9 | 2.2 | — | — | — | — | NA | NA | NA | 6.3 | 4.6 | — | — | — | — |
| Hamsters (8 wk) | |||||||||||||||||||||||||||
| Nasal turb | NA | NA | NA | 4.1 | 4.0 | 4.7 | — | 2.6 | — | — | NA | NA | 6.0 | 5.5 | 5.5 | 2.5 | — | — | — | NA | NA | 6.3 | 6.1 | 5.7 | — | — | — |
| Trachea | NA | NA | NA | 3.6 | 5.0 | 4.0 | — | — | — | — | NA | NA | 4.5 | 4.5 | 4.5 | — | — | — | — | NA | NA | 4.9 | 3.6 | 3.3 | — | — | — |
| Lung | |||||||||||||||||||||||||||
| R cra/acce | NA | NA | NA | — | 3.0 | — | — | — | — | NA | NA | NA | 5.1 | — | — | — | — | — | NA | NA | NA | 5.3 | 6.0 | 3.9 | — | — | — |
| R middle | NA | NA | NA | 2.7 | 3.8 | — | — | — | — | NA | NA | NA | 5.4 | 3.7 | — | — | — | 1.9 | NA | NA | NA | 6.0 | — | — | — | — | — |
| R caudal | NA | NA | NA | 3.1 | 4.4 | — | — | — | — | NA | NA | NA | 5.1 | 5.1 | — | — | — | — | NA | NA | NA | 2.0 | — | — | — | — | — |
| L | NA | NA | NA | 4.3 | 6.2 | — | — | — | — | NA | NA | NA | 5.1 | 2.2 | — | — | — | — | NA | NA | NA | — | — | 3.2 | — | — | — |
Six-week-old female DBA/2 mice and BALB/c mice and 4- or 8-week-old female Syrian hamsters were anesthetized and intranasally inoculated with 106 PFU of the H3N2/2013, H3N2/2014, or H3N2/2016 virus. Three animals per group were euthanized at 24 h postinfection (hpi) (mice only) and on days 3 and 6 postinfection.
Nasal turb, nasal turbinate; R cra/acce, right cranial and accessory lobes; R middle, right middle lobe; R caudal, right caudal lobe; L, left lobe.
—, virus not detected.
NA, not available.
Growth properties of pdmH1N1 viruses in hamsters and mice.
We performed experiments with pandemic H1N1 (pdmH1N1) viruses similar to those performed for H3N2 viruses and described above. Both the pandemic A/Hiroshima/19/2013 (pdmH1N1/2013) and pandemic A/Tokyo/IMS1-1/2014 (pdmH1N1/2014) viruses were pathogenic in mice. Specifically, all of the infected DBA/2 mice died during the observation period (Fig. 3B), and although none of the infected BALB/c mice died, their body weights decreased (Fig. 3B) and virus titers were very high in their respiratory tracts (Table 2). Hamsters were also highly susceptible to the pdmH1N1 viruses. Although neither the 4-week-old hamsters nor the 8-week-old hamsters infected with pdmH1N1/2013 virus showed any body weight loss (Fig. 3B, left), we did observe a decrease in the body weights of the 4- and 8-week-old hamsters infected with pdmH1N1/2014 virus (Fig. 3B, right). Virus was detected in the nasal washes of all of the pdmH1N1/2013- and pdmH1N1/2014-infected hamsters at least until day 4 (Fig. 4B). In addition, high titers of both the pdmH1N1/2013 and pdmH1N1/2014 viruses were detected in the respiratory tracts of the hamsters, especially on day 3 (Table 2). These results indicate that hamsters are highly susceptible to pdmH1N1 viruses.
TABLE 2.
Virus titers in tissues of animals infected with pdmH1N1 virusesa
| Animal (age) and organb | Titer (log10 PFU/g) |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pdmH1N1/2013 |
pdmH1N1/2014 |
|||||||||||||||||
| 24 hpi |
Day 3 |
Day 6 |
24 hpi |
Day 3 |
Day 6 |
|||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
| DBA/2 mice | ||||||||||||||||||
| Nasal turb | 8.3 | 8.3 | 8.2 | 7.5 | 8.3 | 7.1 | NDc | ND | 8.2 | 9.7 | 8.4 | 8.2 | 8.1 | 7.9 | 8.2 | ND | ND | ND |
| Trachea | 8.5 | 8.9 | 8.8 | 7.1 | 7.1 | 7.3 | ND | ND | 7.3 | 8.1 | 7.6 | 8.2 | 7.2 | 7.5 | 7.4 | ND | ND | ND |
| Lung | 8.5 | 8.6 | 8.4 | 7.1 | 7.2 | 7.4 | ND | ND | 7.2 | 9.0 | 9.0 | 9.0 | 7.7 | 8.2 | 8.1 | ND | ND | ND |
| BALB/c mice | ||||||||||||||||||
| Nasal turb | 8.1 | 8.8 | 8.1 | 7.4 | 7.3 | 7.5 | 7.3 | 7.1 | 7.1 | 8.4 | 8.5 | 8.5 | 8.1 | 9.0 | 7.9 | 7.0 | 6.5 | 6.4 |
| Trachea | 7.9 | 8.3 | 8.1 | 6.6 | 6.0 | 6.1 | 5.3 | 6.2 | 6.3 | 6.9 | 7.4 | 7.3 | 5.9 | 5.9 | 5.3 | 5.3 | 5.8 | 4.1 |
| Lung | 8.3 | 8.3 | 8.5 | 7.1 | 7.2 | 6.7 | 5.4 | 5.6 | 5.5 | 8.9 | 8.6 | 8.6 | 7.3 | 7.4 | 6.5 | 4.6 | 4.3 | 4.3 |
| Hamsters (4 wk) | ||||||||||||||||||
| Nasal turb | NAd | NA | NA | 8.2 | 8.5 | 7.8 | 2.8 | 2.8 | 3.4 | NA | NA | NA | 8.8 | 8.7 | 9.0 | 2.9 | 3.1 | 4.5 |
| Trachea | NA | NA | NA | 8.1 | 7.7 | 8.0 | —e | — | 2.3 | NA | NA | NA | 8.4 | 8.9 | 8.5 | — | — | 4.4 |
| Lung | ||||||||||||||||||
| R cra/acce | NA | NA | NA | 8.2 | 6.5 | — | — | 2.8 | 3.0 | NA | NA | NA | 8.2 | 6.5 | 7.5 | 2.1 | 2.9 | 1.8 |
| R middle | NA | NA | NA | 6.3 | 6.4 | — | — | 1.9 | 3.3 | NA | NA | NA | 6.9 | 7.7 | 6.7 | — | 5.8 | 5.8 |
| R caudal | NA | NA | NA | 6.3 | 6.4 | — | — | 2.9 | 4.7 | NA | NA | NA | 8.2 | 7.2 | 8.1 | 3.4 | 4.9 | 2.7 |
| L | NA | NA | NA | 7.4 | 6.1 | — | — | 1.8 | 2.9 | NA | NA | NA | 7.1 | 7.6 | 7.4 | — | 4.4 | — |
| Hamsters (8 wk) | ||||||||||||||||||
| Nasal turb | NA | NA | NA | 8.0 | 7.5 | 7.5 | 5.4 | 5.5 | 5.7 | — | NA | NA | 8.7 | 8.9 | 8.5 | 3.7 | 3.4 | 4.7 |
| Trachea | NA | NA | NA | 7.4 | 6.2 | 7.1 | 2.5 | 2.4 | 1.8 | — | NA | NA | 8.5 | 8.5 | 8.2 | 2.4 | 2.3 | 2.3 |
| Lung | ||||||||||||||||||
| R cra/acce | NA | NA | NA | 5.3 | 7.0 | 7.2 | 3.3 | — | — | NA | NA | NA | 7.2 | 7.4 | 7.6 | 4.6 | — | — |
| R middle | NA | NA | NA | 6.2 | 7.3 | 7.0 | 1.9 | — | 6.3 | NA | NA | NA | 7.5 | 7.1 | 6.8 | 3.0 | 5.9 | 3.6 |
| R caudal | NA | NA | NA | 6.8 | 7.8 | 7.0 | 5.3 | — | 1.6 | NA | NA | NA | 7.4 | 7.3 | 7.3 | 3.3 | 1.6 | — |
| L | NA | NA | NA | 7.0 | 6.7 | 6.9 | 5.3 | 2.2 | 1.9 | NA | NA | NA | 7.2 | 7.3 | 7.1 | 4.6 | 2.4 | 2.0 |
Six-week-old female DBA/2 mice and BALB/c mice and 4- or 8-week-old female Syrian hamsters were anesthetized and intranasally inoculated with 106 PFU of pdmH1N1/2013 or pdmH1N1/2014 virus. Three animals per group were euthanized at 24 h postinfection (hpi) (mice only) and on days 3 and 6 postinfection.
Nasal turb, nasal turbinate; R cra/acce, right cranial and accessory lobes; R middle, right middle lobe; R caudal, right caudal lobe; L, left lobe.
ND, not done. These animals died on day 5 (DBA/2 mice 7, 17, 18, and 19) or day 6 (DBA/2 mouse 8).
NA, not available.
—, virus not detected.
Growth properties of type B viruses in hamsters and mice.
We further tested the susceptibility of mice and hamsters to influenza B viruses as described above for the H3N2 and pdmH1N1 viruses. Both B/Kamakura/8/2014 (Victoria lineage; Victoria/2014) and B/Kamakura/10/2014 (Yamagata lineage; Yamagata/2014) were highly pathogenic in DBA/2 mice. Two of 4 DBA/2 mice infected with each of the viruses died during the observation period (Fig. 3C), but none of the infected BALB/c mice died from their type B virus infection. Body weight loss was observed among the BALB/c mice after Yamagata/2014 infection but not after Victoria/2014 infection (Fig. 3C). In contrast, Victoria/2014 infection blocked the body weight gain of both the 4- and 8-week-old hamsters (Fig. 3C). Although the titers in the Victoria/2014- and the Yamagata/2014-infected hamsters were lower than those in the pdmH1N1 virus-infected hamsters, virus was detected in the nasal washes of both the Victoria/2014- and the Yamagata/2014-infected hamsters at least until day 4 (Fig. 4C). Similarly, although the titers in the respiratory tracts of the type B virus-infected mice were lower than those of the pdmH1N1 viruses, appreciably high Victoria/2014 and Yamagata/2014 virus titers were detected in the respiratory tracts of both the DBA/2 and BALB/c mice (Tables 2 and 3). Also in hamsters, although some variations were found depending on the individual animals, the viruses used, and the age of the animals, both type B viruses replicated appreciably well in the respiratory organs. These results indicate that hamsters are also susceptible to type B viruses.
TABLE 3.
Virus titers in tissues of animals infected with influenza B virusesa
| Animal (age) and organb | Titer (log10 PFU/g) |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Victoria/2014 |
Yamagata/2014 |
|||||||||||||||||
| 24 hpi |
Day 3 |
Day 6 |
24 hpi |
Day 3 |
Day 6 |
|||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
| DBA/2 mice | ||||||||||||||||||
| Nasal turb | 6.7 | 6.5 | 6.6 | 6.0 | 6.1 | 6.0 | 4.8 | 5.2 | 4.1 | 5.8 | 5.3 | 6.1 | 5.4 | 5.2 | 3.9 | 2.6 | 3.1 | 2.6 |
| Trachea | 6.9 | 7.1 | 6.6 | 5.1 | 5.9 | 5.3 | 4.7 | 4.8 | 3.4 | 5.2 | 4.2 | 4.5 | 2.8 | 4.2 | —c | 4.2 | 5.0 | — |
| Lung | 6.4 | 6.4 | 6.6 | 6.2 | 6.1 | 6.2 | 5.2 | 5.9 | 4.7 | 4.7 | 4.7 | 3.6 | 3.0 | 4.4 | 3.4 | 2.3 | 2.5 | — |
| BALB/c mice | ||||||||||||||||||
| Nasal turb | 5.8 | 5.6 | 6.3 | 6.4 | 6.3 | 6.0 | 3.7 | 3.2 | 3.3 | 5.3 | 5.1 | 5.1 | 5.3 | 5.0 | 5.0 | 2.4 | — | 2.6 |
| Trachea | 5.9 | 5.6 | 6.1 | 6.9 | 5.3 | 5.3 | — | — | — | 3.1 | 4.3 | 3.2 | — | 2.9 | — | — | — | — |
| Lung | 5.7 | 5.2 | 5.6 | 5.6 | 5.1 | 4.9 | 1.7 | 1.6 | 1.6 | 5.4 | 5.1 | 5.4 | 3.4 | 3.2 | 3.5 | — | — | — |
| Hamsters (4 wk) | ||||||||||||||||||
| Nasal turb | NAd | NA | NA | 6.7 | 6.5 | 6.5 | 5.2 | — | 2.5 | NA | NA | NA | 7.3 | 7.6 | 7.8 | 2.6 | 2.4 | 2.0 |
| Trachea | NA | NA | NA | 5.9 | 5.9 | 6.0 | — | — | 2.8 | NA | NA | NA | 3.4 | 5.6 | 5.7 | — | — | — |
| Lung | ||||||||||||||||||
| R cra/acce | NA | NA | NA | 6.5 | 6.8 | — | — | — | 2.8 | NA | NA | NA | 2.9 | 3.1 | 5.6 | — | — | — |
| R middle | NA | NA | NA | 6.8 | 6.6 | — | — | — | 3.2 | NA | NA | NA | — | — | 6.3 | — | — | — |
| R caudal | NA | NA | NA | 6.5 | 7.0 | — | — | — | 3.0 | NA | NA | NA | 2.7 | 2.7 | 6.2 | — | — | — |
| L | NA | NA | NA | 6.6 | 6.7 | — | — | — | 4.2 | NA | NA | NA | — | 5.4 | 6.5 | — | — | — |
| Hamsters (8 wk) | ||||||||||||||||||
| Nasal turb | NA | NA | NA | 6.7 | 6.9 | 6.1 | 4.3 | 3.0 | 3.3 | — | NA | NA | 6.7 | 7.9 | 6.9 | 3.0 | 2.6 | 3.1 |
| Trachea | NA | NA | NA | 5.6 | 6.2 | 5.7 | — | 2.2 | — | — | NA | NA | 4.2 | 6.0 | 6.1 | — | — | — |
| Lung | ||||||||||||||||||
| R cra/acce | NA | NA | NA | 6.6 | 3.3 | 2.8 | 4.4 | — | — | NA | NA | NA | — | — | 6.4 | — | — | — |
| R middle | NA | NA | NA | 6.8 | 3.1 | 2.6 | 4.8 | — | — | NA | NA | NA | — | — | 4.8 | — | — | — |
| R caudal | NA | NA | NA | 7.0 | 2.1 | 3.1 | 2.0 | — | — | NA | NA | NA | 2.9 | — | 6.6 | — | — | — |
| L | NA | NA | NA | 6.9 | 3.0 | 2.8 | — | 2.1 | 2.6 | NA | NA | NA | — | 1.7 | 3.7 | — | — | — |
Six-week-old female DBA/2 mice and BALB/c mice and 4- or 8-week-old female Syrian hamsters were anesthetized and intranasally inoculated with 106 PFU of Victoria/2014 or Yamagata/2014 virus. Three animals per group were euthanized at 24 h postinfection (hpi) (mice only) and on days 3 and 6 postinfection.
Nasal turb, nasal turbinate; R cra/acce, right cranial and accessory lobes; R middle, right middle lobe; R caudal, right caudal lobe; L, left lobe.
—, virus not detected.
NA, not available.
Pathological analyses of influenza virus-infected animals.
Four-week-old female hamsters and 6-week-old female BALB/c mice or DBA/2 mice were infected with H3N2/2013 (n = 4), pdmH1N1/2013 (n = 4), or Victoria/2014 (n = 4) virus. On days 3 (n = 2) and 6 (n = 2) postinfection, the animals were euthanized and their organs were collected for pathological examinations. The pdmH1N1/2013 virus-infected DBA/2 mice scheduled for sampling on day 6 died on day 3 and on day 6. The DBA/2 mouse that died on day 6 was dissected just after death for pathological analyses.
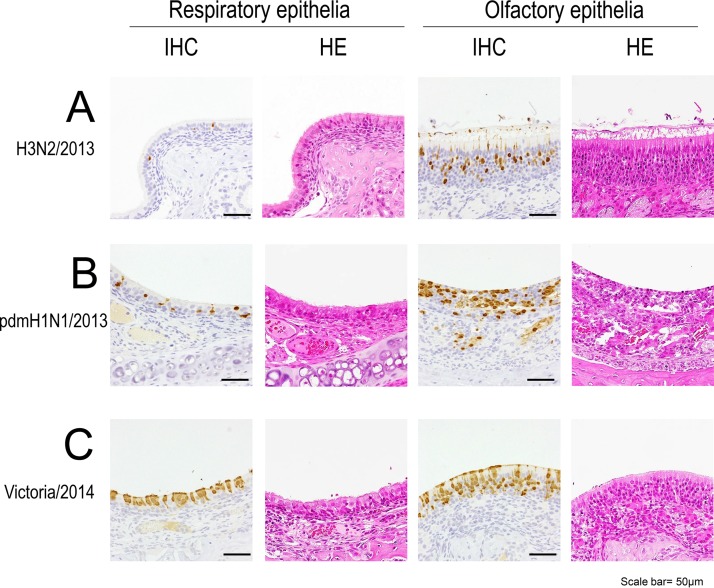
The number of antigen-positive cells detected by immunohistochemistry (Table 4) showed a pattern similar to that of the virus titers (Tables 1 to 3). In the case of H3N2/2013 virus infection, virus antigens were not detected in any DBA/2 or BALB/c mouse but were detected in the nasal turbinate of one hamster and in the trachea and bronchus of another hamster (Table 4). There were few antigen-positive cells, and it was difficult to determine the cell tropism of the H3N2 viruses. Histopathological changes were limited in the nasal turbinate of H3N2/2013-infected hamsters, and virus antigens were mainly detected in the olfactory epithelia (Fig. 5). In contrast, in the cases of pdmH1N1/2013 or Victoria/2014 virus infection, virus antigens were detected in the respiratory organs of all animals infected with either pdmH1N1/2013 virus or Victoria/2014 virus (Table 4). In the nasal turbinate of the infected hamsters, inflammatory cells infiltrated the lamina propria (Fig. 5). The olfactory epithelia of the pdmH1N1/2013-infected hamsters were partially eroded (Fig. 5). There were fewer antigen-positive cells in the hamsters than in the mice (Table 4). The distribution of virus antigens in the nasal turbinate differed between type A (H3N2 and pdmH1N1/2013)- and type B (Victoria/2014)-infected hamsters. In the nasal turbinate of the H3N2- or pdmH1N1/2013-infected hamsters, virus antigens were detected mainly in the olfactory epithelia rather than in the respiratory epithelia (Fig. 5). In contrast, in the nasal turbinate of the Victoria/2014-infected hamsters, virus antigens were detected in both the respiratory epithelia and the olfactory epithelia (Fig. 5).
TABLE 4.
Number of antigen-positive cells in H3N2/2013-, pdmH1N1/2013-, and Victoria/2014-infected animalsa
| Animal and organ | No. of antigen-positive cellsb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H3N2/2013 |
pdmH1N1/2013 |
Victoria/2014 |
||||||||||
| Day 3 |
Day 6 |
Day 3 |
Day 6 |
Day 3 |
Day 6 |
|||||||
| 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | |
| DBA/2 mice | ||||||||||||
| Nasal turbinate | − | − | − | − | + | + | + | NDc | ++ | ++ | + | + |
| Trachea | − | − | − | − | + | + | + | ND | ++ | NAd | + | + |
| Bronchus | − | − | − | − | ++ | ++ | + | ND | ++ | ++ | + | ++ |
| Alveolus | − | − | − | − | ++ | ++ | ++ | ND | + | ++ | + | ++ |
| BALB/c mice | ||||||||||||
| Nasal turbinate | − | − | − | − | ++ | ++ | + | − | ++ | ++ | + | +/− |
| Trachea | − | − | − | − | + | + | + | +/− | ++ | ++ | + | +/− |
| Bronchus | − | − | − | − | ++ | + | + | + | ++ | ++ | + | + |
| Alveolus | − | − | − | − | ++ | + | + | + | ++ | + | + | + |
| Syrian hamster (4 wk) | ||||||||||||
| Nasal turbinate | − | + | − | − | + | + | +/− | + | ++ | ++ | + | + |
| Trachea | + | − | − | − | + | + | − | − | + | + | +/− | +/− |
| Bronchus | + | − | − | − | + | + | − | − | + | + | +/− | +/− |
| Alveolus | − | − | − | − | +/− | +/− | − | − | +/− | +/− | +/− | +/− |
Six-week-old female DBA/2 mice and BALB/c mice and 4-week-old female Syrian hamsters were anesthetized and intranasally inoculated with 106 PFU of H3N2/2013, pdmH1N1/2013, or Victoria/2014 virus. Two animals per group were euthanized on days 3 and 6 postinfection.
−, no antigen-positive cells; +/−, less than 5 antigen-positive cells; +, more than 6 antigen-positive cells; ++, widespread antigen-positive cells.
ND, not done. This animal died on day 3 postinfection.
NA, not available.
FIG 5.
Pathological examination of the respiratory epithelia (left) and olfactory epithelia (right) of the nasal turbinates of infected 4-week-old hamsters. The images show the nasal turbinates of hamsters on day 3 postinfection with 106 PFU of H3N2/2013 (A), pdmH1N1/2013 (B), or Victoria/2014 (C) virus. HE, hematoxylin and eosin staining; IHC, immunohistochemistry for the detection of influenza virus NP antigen.
Transmissibility of influenza viruses in hamsters.
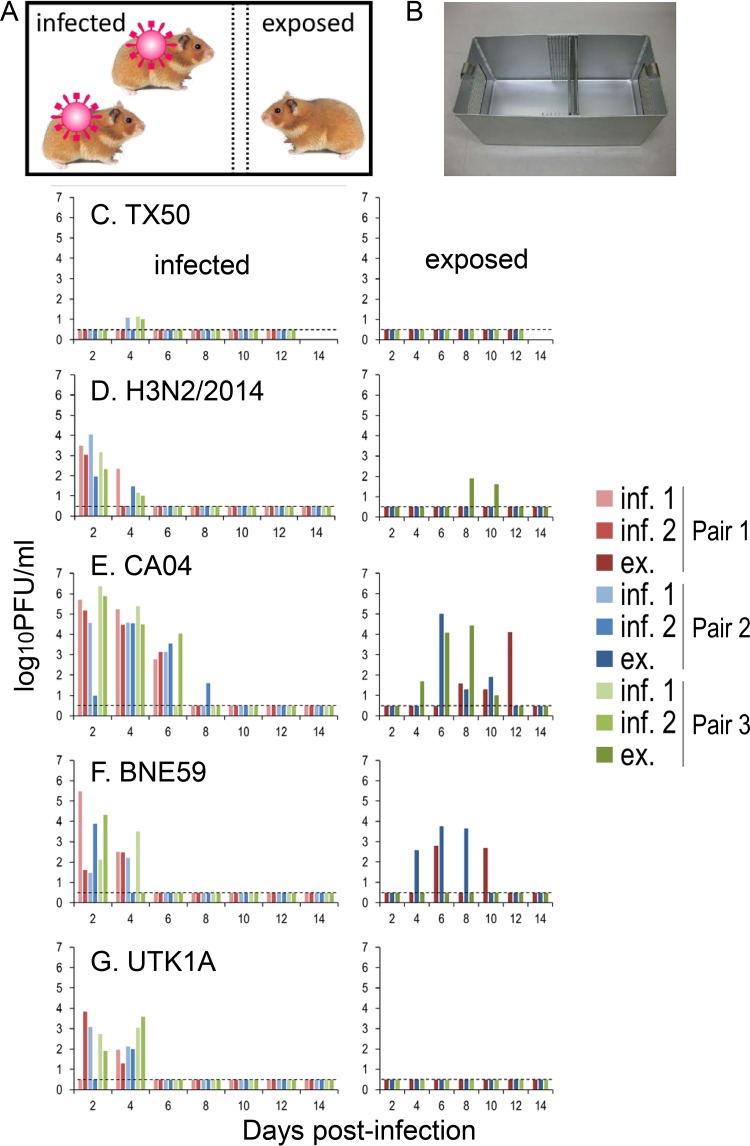
To assess the transmissibility of influenza viruses in hamsters, two animals each infected with 106 PFU of A/Texas/50/2012 (H3N2; TX50), H3N2/2014, A/California/04/2009 (pdmH1N1; CA04), A/Brisbane/59/2007 (H1N1; BNE59), or B/Yokohama/UT-K1A/2011 (type B Victoria linage; UTK1A) were placed in the larger room of a transmission cage, and on the next day, a naive hamster was placed in the adjacent smaller room of the cage (Fig. 6A). Three sets of hamsters (nine animals in total) were used for each virus. We recovered viruses from the nasal washes of all infected hamsters, except for three animals infected with TX50 (Fig. 6C to G, left). No virus was detected in the nasal washes of all three hamsters that were exposed to hamsters infected with TX50 (H3N2) or UTK1A (type B) (Fig. 6C and G). In contrast, all of the hamsters that were exposed to hamsters infected with CA04 (pdmH1N1) (Fig. 6E), two of three hamsters (pairs 1 and 2) that were exposed to hamsters infected with BNE59 (H1N1) (Fig. 6F), and one of three hamsters (pair 3) that were exposed to hamsters infected with H3N2/2014(H3N2) (Fig. 6D) shed viruses. Serum antibody titers against each virus confirmed infection of the animals from which virus was recovered, whereas the exposed hamsters from which virus was not recovered did not seroconvert, with the exception of hamsters exposed to hamsters infected with H3N2/2014(H3N2) (data not shown); all three hamsters exposed to the H3N2/2014(H3N2)-infected group seroconverted, with the virus neutralization titers being 1:16 for pair 1, 1:256 for pair 2, and 1:128 for pair 3, indicating that this virus transmitted to all three hamsters. These results indicate that hamsters can be used to evaluate the airborne transmissibility of human influenza viruses.
FIG 6.
Respiratory droplet transmission of influenza viruses in hamsters. (A, B) Schematic representation (A) and photograph (B) of the transmission cage used for the hamster transmission studies. This cage has two wire-mesh partitions that prevent direct and indirect contact between the animals but allow the spread of influenza virus through the air. (C to G) Three groups of hamsters (two per group) were inoculated intranasally with 106 PFU of TX50 (C), H3N2/2014 (D), CA04 (E), BNE59 (F), or UTK1A (G) virus and then placed in the larger room of a transmission cage (day 0). At 24 h after infection (day 1), one naive exposed hamster per group was placed in the adjacent smaller room (A). Nasal washes were collected every other day from both infected (C to G, left) and exposed (C to G, right) animals for virus titration. Virus titers were determined by using a plaque assay on MDCK cells. The lower limit of detection is indicated by the horizontal dashed lines. inf., infected hamster; ex., exposed hamster.
DISCUSSION
In this study, we demonstrated that hamsters are susceptible to influenza viruses, including the recent H3N2 viruses. Although hamsters did not show weight loss or clinical signs of H3N2 virus infection, we detected virus antigens in the respiratory tracts of infected hamsters without adaptation of the viruses. Hamsters are easier to handle than ferrets, and recent H3N2 viruses do not appreciably replicate in mice; therefore, these findings indicate that hamsters may represent an alternative rodent model for studies of recent human influenza viruses, especially H3N2 viruses.
The distribution of sialic acids on the epithelial cells of the respiratory tract of ferrets (38) is similar to that on the epithelial cells of the respiratory tract of humans, in that SAα2,6Gal is dominant in the respiratory tract and SAα2,3Gal is expressed at low levels in the lower respiratory tract (39). In contrast, SAα2,3Gal is expressed in the respiratory tract of C57BL/6J mice, but SAα2,6Gal is not (40). These differences in receptor distribution might play a role in the differences in sensitivity to influenza viruses among animal species. Interestingly, the viruses tested in this study showed different cell tropisms. The type B viruses infected both olfactory epithelia and respiratory epithelia, but the H3N2 and pdmH1N1 viruses preferentially infected the olfactory epithelia (Fig. 5). The olfactory epithelial cells reacted with MAA II, which recognizes SAα2,3Gal, but not with SNA I, which recognizes SAα2,6Gal (Fig. 1B and D). Clinical human influenza viruses isolated in Madin-Darby canine kidney (MDCK) cells preferentially bind to SAα2,6Gal (1). Therefore, the distribution of these types of sialyloligosaccharides, as determined with MAA II and SNA I lectins, is not consistent with the receptor specificity of the viruses used. It may be that the SAα2,6Gal that is present in the hamster olfactory epithelia is not detectable with SNA I. Further studies are needed to test this possibility.
We also found that H1N1 and H3N2 viruses, but not type B viruses, are transmissible by the airborne route in hamsters (Fig. 6). Considering the difference in the virus titers in the nasal turbinates among these viruses, the transmissibility of influenza viruses in hamsters may depend on the virus titers in the upper respiratory tract. Although it is important to test whether the viruses can further transmit to other naive animals from exposed animals, we are currently unable to perform such an experiment due to the moratorium on gain-of-function experiments. Although the titers of type B virus in the nasal wash specimens were not particularly low, this virus was not transmissible by the airborne route (Fig. 6G). Nevertheless, our data suggest that hamsters may represent a useful model of transmission of influenza viruses. For evaluation of the airborne transmissibility of different influenza viruses, ferrets have been used extensively and guinea pigs have been used by some groups. It is important to compare these animal models side by side with the hamster model when evaluating the transmissibility of influenza viruses and its determinants.
In conclusion, hamsters have the potential to be a useful small-animal model for studies of influenza virus infection. They can be used for pathogenicity studies of not only the recent H3N2 viruses but also other strains. Moreover, they can also be used for studies of the transmission of some virus strains.
MATERIALS AND METHODS
Cells and viruses.
MDCK cells were maintained in Eagle's minimal essential medium (MEM) containing 5% newborn calf serum at 37°C in 5% CO2. For infectivity studies, we used three A(H3N2) viruses (A/Tokyo/IMS6-1/2013 [H3N2/2013], A/Tokyo/IMS2-1/2014 [H3N2/2014], and A/Tokyo/UT-HP002/2016 [H3N2/2016]), two A(H1N1)pdm09 viruses (A/Hiroshima/19/2013 [pdmH1N1/2013] and A/Tokyo/IMS1-1/2014 [pdmH1N1/2014]), and two type B viruses (a Victoria lineage virus, B/Kamakura/8/2014 [Victoria/2014], and a Yamagata lineage virus, B/Kamakura/10/2014 [Yamagata/2014]). For transmission studies, we used A/Texas/50/2012 (H3N2; TX50), A/Tokyo/IMS2-1/2014 (H3N2/2014), A/California/04/2009 (pdmH1N1; CA04), A/Brisbane/59/2007 (H1N1; BNE59), and B/Yokohama/UT-K1A/2011 (type B Victoria linage; UTK1A). All viruses except for BNE59 were isolated in MDCK cells or AX-4 cells (AX-4 cells are derivatives of MDCK cells expressing a larger amount of SAα2,6Gal) and then propagated them in MDCK cells. BNE59 was obtained from the U.S. CDC; it had been propagated in the allantoic cavity of embryonated chicken eggs.
Plaque assay.
Viruses were diluted in MEM containing 0.3% bovine serum albumin (BSA). Confluent monolayers of MDCK cells were washed with MEM containing 0.3% BSA, infected with diluted viruses, and incubated for 30 to 60 min at 37°C. After the virus inoculum was removed, the cells were washed with MEM containing 0.3% BSA and overlaid with a 1:1 mixture of 2× MEM–0.6% BSA and 2% agarose containing 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin. The plates were incubated at 37°C for 48 h before virus plaques were counted.
Experimental infection.
Six-week-old female BALB/c mice and DBA/2 mice and 4- or 8-week-old female Syrian hamsters (Japan SLC Inc., Shizuoka, Japan) were used for this study. The animal room was keep at 25°C and 50% humidity. Four mice and three hamsters per group were anesthetized with isoflurane and intranasally inoculated with 106 PFU/animal (50 μl for mice and 100 μl for hamsters) of H3N2/2013, H3N2/2014, H3N2/2016, pdmH1N1/2013, pdmH1N1/2014, Victoria/2014, or Yamagata/2014 viruses. Body weight and survival were monitored daily for 10 to 14 days postinfection (dpi). Baseline body weights were measured prior to infection. Nasal wash specimens were collected from each hamster with 400 μl of phosphate-buffered saline (PBS) every other day for virus titration. To assess virus growth in the respiratory organs, three mice or three hamsters per group were infected intranasally with 106 PFU of viruses and euthanized, and nasal turbinates, tracheas, and lungs were collected on days 1 (only for mice), 3, and 6 postinfection. The collected organs were homogenized with MEM containing 0.3% BSA and titrated in MDCK cells by using plaque assays.
Pathological examination.
The excised respiratory tract tissues were fixed in 4% paraformaldehyde phosphate (PFA) buffer solution for 48 h and then processed for paraffin embedding. Nasal samples were immersed in EDTA solution for decalcification, after being fixed in PFA. The paraffin blocks were cut into 3-μm-thick sections and were mounted on silane-coated glass slides. To detect SAα2,6Gal and SAα2,3Gal, the sections were pretreated with 0.05% trypsin (Difco Laboratories, Detroit, MI, USA) at 37°C for 15 min and 0.3% hydrogen peroxide at room temperature for 30 min. They were then incubated at 4°C overnight with biotin-conjugated SNA I (EY Laboratories) for SAα2,6Gal detection and biotinylated conjugated MAA II (Vector Laboratories) for SAα2,3Gal detection. After being washed, the sections were then incubated with horseradish peroxidase-conjugated streptavidin and were visualized by staining with 3,3′-diaminobenzidine (DAB). The sections were also stained using a standard hematoxylin and eosin procedure, and each serial section was processed for immunohistological staining with a rabbit polyclonal antibody for type A influenza virus nucleoprotein and a mouse polyclonal antibody for type B influenza virus (prepared in the Department of Pathology, National Institute of Infectious Diseases, Tokyo, Japan). Specific antigen-antibody reactions were visualized with DAB staining by using a Dako Envision system (Dako Cytomation).
Airborne transmission study.
For transmission studies in hamsters, animals were housed in transmission cages that had two wire-mesh partitions that prevented direct and indirect contact between animals but allowed the spread of influenza virus through the air (Showa Science) (Fig. 6A and B). Paper chips (Paper Clean; Japan SLC Inc.) were used for bedding to prevent the production of micropowder. The animal room was keep at 25°C and 50% humidity. Two 8-week-old hamsters were inoculated intranasally with 106 PFU (100 μl) of virus and placed in the larger room of the transmission cage (Fig. 6A) (day 0). At 24 h after infection (day 1), one naive 8-week-old hamster was placed in the smaller room adjacent to the inoculated hamsters. Three sets of hamsters (i.e., nine animals) were used for each virus tested. The hamsters were monitored for changes in body weight and the presence of clinical signs. To assess viral replication in nasal turbinates, we determined the virus titers in the nasal wash specimens collected from virus-inoculated and virus-exposed hamsters on day 2 after inoculation and then every other day.
Ethics statements.
Our research protocol for the animal studies is in accordance with the Regulations for Animal Care at the University of Tokyo, Tokyo, Japan, and was approved by the Animal Experiment Committee of the Institute of Medical Science, the University of Tokyo (approval numbers PA14-35 and PA15-10).
ACKNOWLEDGMENTS
We thank Susan Watson for editing the manuscript and Yuko Sato, Tomohiko Koibuchi, Michiko Koga, Eisuke Adachi, Tadashi Kikuchi, Hirofumi Kobayashi, Ryuta Uraki, and Maki Kiso for technical support.
This research was supported by Leading Advanced Projects for Medical Innovation (LEAP) from the Japan Agency for Medical Research and Development (AMED), by Grants-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports, and Technology (MEXT) of Japan (no. 16H06429, 16K21723, and 16H06434), and by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from MEXT and AMED.
REFERENCES
- 1.Wright PF, Neumann G, Kawaoka Y. 2013. Orthomyxoviruses, p 1186–1243. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.O'Donnell CD, Subbarao K. 2011. The contribution of animal models to the understanding of the host range and virulence of influenza A viruses. Microbes Infect 13:502–515. doi: 10.1016/j.micinf.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jayaraman A, Chandrasekaran A, Viswanathan K, Raman R, Fox JG, Sasisekharan R. 2012. Decoding the distribution of glycan receptors for human-adapted influenza A viruses in ferret respiratory tract. PLoS One 7:e27517. doi: 10.1371/journal.pone.0027517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belser JA, Katz JM, Tumpey TM. 2011. The ferret as a model organism to study influenza A virus infection. Dis Model Mech 4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carolan LA, Rockman S, Borg K, Guarnaccia T, Reading P, Mosse J, Kelso A, Barr I, Laurie KL. 2015. Characterization of the localized immune response in the respiratory tract of ferrets following infection with influenza A and B viruses. J Virol 90:2838–2848. doi: 10.1128/JVI.02797-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Vries RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y. 2013. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501:551–555. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley AJ, Kawaoka Y. 1999. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol 73:3184–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 11.Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, Sakabe S, Imai M, Hatta Y, Watanabe S, Li C, Yamada S, Fujii K, Murakami S, Imai H, Kakugawa S, Ito M, Takano R, Iwatsuki-Horimoto K, Shimojima M, Horimoto T, Goto H, Takahashi K, Makino A, Ishigaki H, Nakayama M, Okamatsu M, Takahashi K, Warshauer D, Shult PA, Saito R, Suzuki H, Furuta Y, Yamashita M, Mitamura K, Nakano K, Nakamura M, Brockman-Schneider R, Mitamura H, Yamazaki M, Sugaya N, Suresh M, Ozawa M, Neumann G, Gern J, Kida H, et al. . 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. 2013. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501:556–559. doi: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mok CK, Lee HH, Chan MC, Sia SF, Lestra M, Nicholls JM, Zhu H, Guan Y, Peiris JM. 2013. Pathogenicity of the novel A/H7N9 influenza virus in mice. mBio 4:e00362-13. doi: 10.1128/mBio.00362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Bao L, Deng W, Zhu H, Chen T, Lv Q, Li F, Yuan J, Xiang Z, Gao K, Xu Y, Huang L, Li Y, Liu J, Yao Y, Yu P, Yong W, Wei Q, Zhang L, Qin C. 2013. The mouse and ferret models for studying the novel avian-origin human influenza A (H7N9) virus. Virol J 10:253. doi: 10.1186/1743-422X-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniels MJ, Selgrade MK, Doerfler D, Gilmour MI. 2003. Kinetic profile of influenza virus infection in three rat strains. Comp Med 53:293–298. [PubMed] [Google Scholar]
- 16.Lowen AC, Mubareka S, Tumpey TM, Garcia-Sastre A, Palese P. 2006. The guinea pig as a transmission model for human influenza viruses. Proc Natl Acad Sci U S A 103:9988–9992. doi: 10.1073/pnas.0604157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Hoeven N, Belser JA, Szretter KJ, Zeng H, Staeheli P, Swayne DE, Katz JM, Tumpey TM. 2009. Pathogenesis of 1918 pandemic and H5N1 influenza virus infections in a guinea pig model: antiviral potential of exogenous alpha interferon to reduce virus shedding. J Virol 83:2851–2861. doi: 10.1128/JVI.02174-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanco JC, Boukhvalova MS, Perez DR, Vogel SN, Kajon A. 2014. Modeling human respiratory viral infections in the cotton rat (Sigmodon hispidus). J Antivir Antiretrovir 6:40–42. doi: 10.4172/jaa.1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ottolini MG, Blanco JC, Eichelberger MC, Porter DD, Pletneva L, Richardson JY, Prince GA. 2005. The cotton rat provides a useful small-animal model for the study of influenza virus pathogenesis. J Gen Virol 86:2823–2830. doi: 10.1099/vir.0.81145-0. [DOI] [PubMed] [Google Scholar]
- 20.Eichelberger MC. 2007. The cotton rat as a model to study influenza pathogenesis and immunity. Viral Immunol 20:243–249. doi: 10.1089/vim.2007.0017. [DOI] [PubMed] [Google Scholar]
- 21.Boukhvalova MS, Prince GA, Blanco JC. 2009. The cotton rat model of respiratory viral infections. Biologicals 37:152–159. doi: 10.1016/j.biologicals.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanco JC, Pletneva LM, Wan H, Araya Y, Angel M, Oue RO, Sutton TC, Perez DR. 2013. Receptor characterization and susceptibility of cotton rats to avian and 2009 pandemic influenza virus strains. J Virol 87:2036–2045. doi: 10.1128/JVI.00638-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wirotesangthong M, Nagai T, Yamada H, Amnuoypol S, Mungmee C. 2009. Effects of Clinacanthus siamensis leaf extract on influenza virus infection. Microbiol Immunol 53:66–74. doi: 10.1111/j.1348-0421.2008.00095.x. [DOI] [PubMed] [Google Scholar]
- 24.Rudneva IA, Kaverin NV, Varich NL, Gitelman AK, Makhov AM, Klimenko SM, Zhdanov VM. 1986. Studies on the genetic determinants of influenza virus pathogenicity for mice with the use of reassortants between mouse-adapted and non-adapted variants of the same virus strain. Arch Virol 90:237–248. doi: 10.1007/BF01317373. [DOI] [PubMed] [Google Scholar]
- 25.Narasaraju T, Sim MK, Ng HH, Phoon MC, Shanker N, Lal SK, Chow VT. 2009. Adaptation of human influenza H3N2 virus in a mouse pneumonitis model: insights into viral virulence, tissue tropism and host pathogenesis. Microbes Infect 11:2–11. doi: 10.1016/j.micinf.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Ping J, Dankar SK, Forbes NE, Keleta L, Zhou Y, Tyler S, Brown EG. 2010. PB2 and hemagglutinin mutations are major determinants of host range and virulence in mouse-adapted influenza A virus. J Virol 84:10606–10618. doi: 10.1128/JVI.01187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reading PC, Morey LS, Crouch EC, Anders EM. 1997. Collectin-mediated antiviral host defense of the lung: evidence from influenza virus infection of mice. J Virol 71:8204–8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potter CW, Jennings R. 1976. The hamster as a model system for the study of influenza vaccines. Postgrad Med J 52:345–351. doi: 10.1136/pgmj.52.608.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jennings R, Denton MD, Potter CW. 1976. The hamster as an experimental animal for the study of influenza. I. The role of antibody in protection. Med Microbiol Immunol 162:217–226. doi: 10.1007/BF02121000. [DOI] [PubMed] [Google Scholar]
- 30.Jennings R, Phair JP, Denton MD, Potter CW. 1976. The hamster as an experimental animal for the study of influenza. II. The role of spleen cells in protection. Med Microbiol Immunol 162:227–237. doi: 10.1007/BF02121001. [DOI] [PubMed] [Google Scholar]
- 31.Reeve P. 1978. Growth of some attenuated influenza A viruses in hamsters. Med Microbiol Immunol 166:133–139. doi: 10.1007/BF02121142. [DOI] [PubMed] [Google Scholar]
- 32.Reeve P, Pibermann M, Gerendas B. 1981. Studies with some influenza B viruses in cell cultures, hamsters and hamster tracheal organ cultures. Med Microbiol Immunol 169:179–186. doi: 10.1007/BF02123591. [DOI] [PubMed] [Google Scholar]
- 33.Wood JM, Oxford JS, Dunleavy U, Newman RW, Major D, Robertson JS. 1989. Influenza A (H1N1) vaccine efficacy in animal models is influenced by two amino acid substitutions in the hemagglutinin molecule. Virology 171:214–221. doi: 10.1016/0042-6822(89)90528-X. [DOI] [PubMed] [Google Scholar]
- 34.Ali MJ, Teh CZ, Jennings R, Potter CW. 1982. Transmissibility of influenza viruses in hamsters. Arch Virol 72:187–197. doi: 10.1007/BF01348964. [DOI] [PubMed] [Google Scholar]
- 35.Harkema JR, Carey SA, Wagner JG. 2006. The nose revisited: a brief review of the comparative structure, function, and toxicologic pathology of the nasal epithelium. Toxicol Pathol 34:252–269. doi: 10.1080/01926230600713475. [DOI] [PubMed] [Google Scholar]
- 36.Corps KN, Islam Z, Pestka JJ, Harkema JR. 2010. Neurotoxic, inflammatory, and mucosecretory responses in the nasal airways of mice repeatedly exposed to the macrocyclic trichothecene mycotoxin roridin A: dose-response and persistence of injury. Toxicol Pathol 38:429–451. doi: 10.1177/0192623310364026. [DOI] [PubMed] [Google Scholar]
- 37.Pica N, Iyer A, Ramos I, Bouvier NM, Fernandez-Sesma A, Garcia-Sastre A, Lowen AC, Palese P, Steel J. 2011. The DBA.2 mouse is susceptible to disease following infection with a broad, but limited, range of influenza A and B viruses. J Virol 85:12825–12829. doi: 10.1128/JVI.05930-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Q, Wang W, Cheng X, Zengel J, Jin H. 2010. Influenza H1N1 A/Solomon Island/3/06 virus receptor binding specificity correlates with virus pathogenicity, antigenicity, and immunogenicity in ferrets. J Virol 84:4936–4945. doi: 10.1128/JVI.02489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 40.Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL. 2006. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol 80:7469–7480. doi: 10.1128/JVI.02677-05. [DOI] [PMC free article] [PubMed] [Google Scholar]