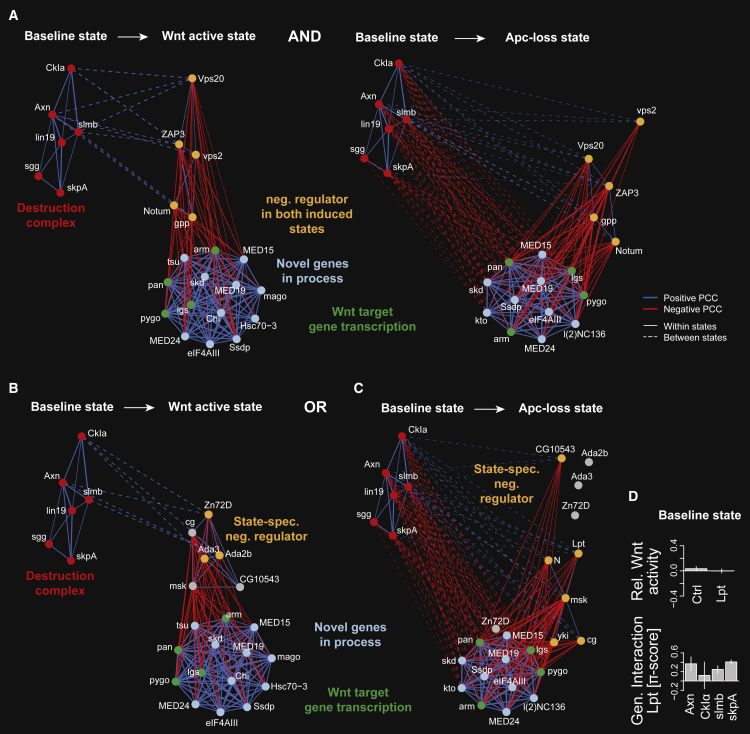
Figure 6.
Genes Resuming a Destruction Complex-like Role in Wnt Signaling in Different Pathway Activation Modes
(A) Putative inhibitors of the Wnt target genes transcription machinery after ligand and Apc-loss-mediated pathway activation.
(B and C) Putative inhibitors of the Wnt target gene transcription machinery exclusively after ligand or Apc-loss-mediated pathway activation. “Bait” genes required for target gene transcription (green nodes) were selected and potentially novel components (light-blue nodes) were called if they shared a CSI > 0.9 in both active pathway states (A) or one but not the other active state (B and C). Potential negative regulators (orange nodes) were added if they shared a between-state similarity (PCC > 0.5) with the destruction complex (red nodes) and a state-specific dissimilarity (PCC < −0.65) with at least one “bait” gene. Genes were connected if their state-specific profiles shared an absolute PCC > 0.65 or if their between-state shared an absolute PCC > 0.5 (dashed lines). Positive and negative correlations are shown as blue and red lines, respectively. Per state, the modules of positive regulators (green, light blue), negative regulators (orange), and the destruction complex (red) were separately arranged using a force-directed layout algorithm. The edge width represents the absolute PCC.
(D) Lpt knockdown effect on Wnt signaling activity (upper panel) and the genetic interaction of Lpt with destruction complex components (lower panel) in the baseline state. The single knockdown phenotype was estimated from phenotypes of two independent dsRNAs in 144 different genetic backgrounds each (error bars show SE of median determined by bootstrapping). Error bars of the genetic interactions show the median absolute deviation of four independent genetic interaction scores for each gene pair.