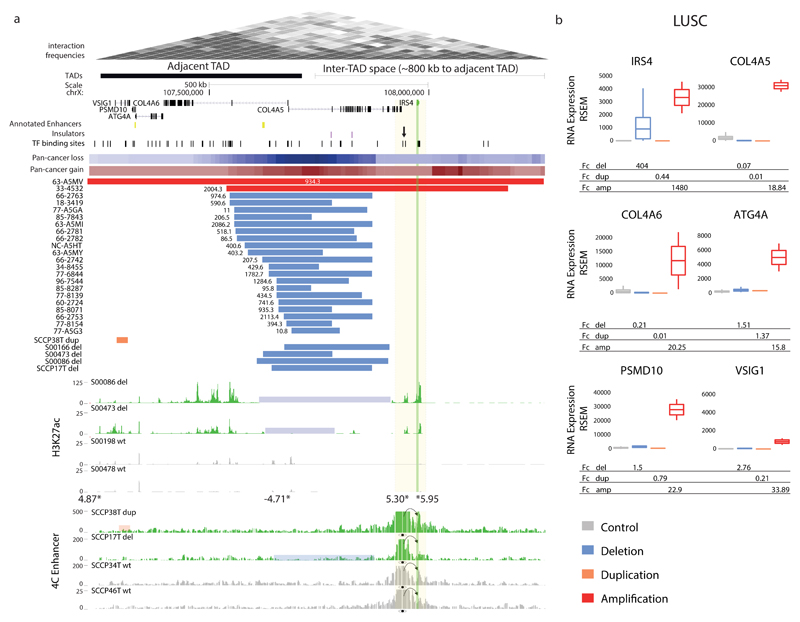
Figure 3. Recurrent SCNAs in cis associate with marked IRS4 expression increase.
(a) Recurrent deletions at a TAD boundary near IRS4, and IRS4 amplifications, associate with IRS4 dysregulation in LUSC. A region near IRS4 exhibiting clustered transcription factor (TF) binding sites (candidate CRE) is highlighted with an arrow. The recurrent deletions were evident both in male and female samples (indicating that both hemizygous and complete losses result in IRS4 overexpression). Summarized SCNAs across cancer types (pan-cancer copy-number gains and losses) shown as heatmaps. The full list of pan-cancer SCNAs at the locus is in Supplementary Table 2. Deletion-carrier samples (del, highlighted in blue) exhibited marked H3K27ac62 at the IRS4 promoter and adjacent candidate CRE. SCNA carrier samples in which chromatin analyses were performed were confirmed to exhibit outlier expression using semi-quantitative RT-PCR and qPCR (Supplementary Fig. 3, Supplementary Table 3, and data not shown). Asterisks depict differentially occupied peaks identified by genome-wide H3K27ac analysis (values adjacent to asterisks show Fc in H3K27ac signal for deletion-carriers vs. non-carriers). Lastly, 4C-Seq experiments using the candidate CRE as a viewpoint in carrier versus non-carrier samples are depicted. dup, duplication; WT, wild-type locus. (b) LUSC expression measurements (unadjusted RSEM gene expression values) for carriers versus non-carriers, revealing IRS4 as the most plausible target. IRS4 expression analyses revealed ~400-fold upregulation in deletion-carriers and >1000-fold for gene amplification carriers (number of control=470; del=24; dup=1; amp=2).