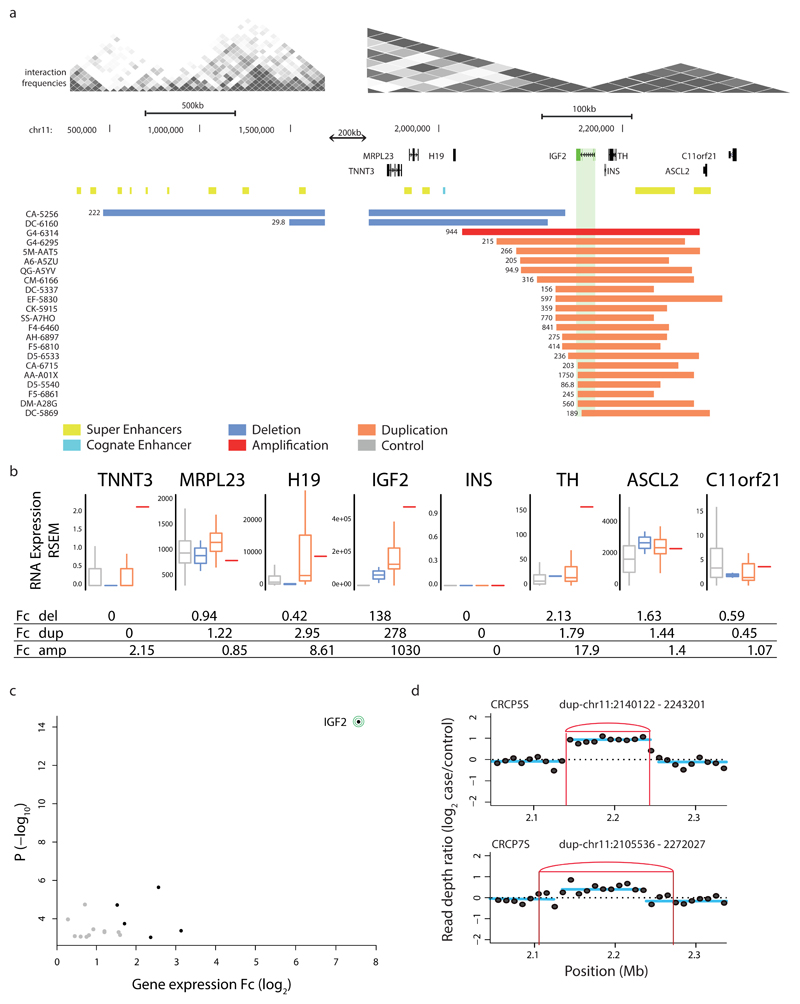
Figure 4. SCNAs associating with marked IGF2 locus overexpression in cis in CRC.
(a) Recurrent somatic duplications at the IGF2 locus (green) associating with IGF2 overexpression encompass a TAD boundary and a super enhancer (yellow) in the adjacent TAD, but do not encompass the known IGF2 cognate enhancer (light blue). Somatic deletions in cis extend over additional TAD boundaries. (b) Boxplots depict expression-SCNAs relationships for all protein-coding genes within the respective TAD, with IGF2 showing the by far most marked relationship making it the most likely target of these recurrent SCNAs in cis (boxplots separating into del carriers, dup carriers, amplification (amp; >4 copies) carriers, and control samples lacking SCNAs in cis). (c) Volcano plot of CESAM hits in CRC, with nominal P-values plotted versus log2-expression change based on all samples with SCNAs in TAD (CESAM hits are depicted in black; IGF2 is highlighted). (d) Structural variant detection by long-insert size paired-end sequencing4, followed by DELLY276 analysis, identified presence of TAD-spanning IGF2 locus tandem duplication in spheroid samples CRCP5S and CRCP7S (IGF2 outlier expression was verified in both samples by qPCR; see Supplementary Fig. 13, Supplementary Table 4).