Figure 3.
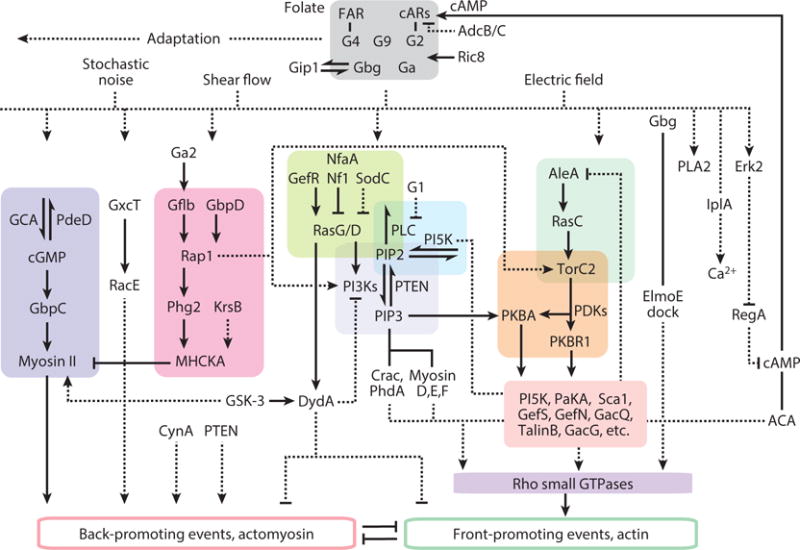
Network of signal transduction pathways involved in directed cell migration in Dictyostelium. Architecture is based on biochemical and genetic analysis and reflects an extensive series of experiments by independent investigators. In most cases connections represent interpretations of phenotypes or stimulus-induced biochemical and biosensor behavior, contrasted to wild type, in cells carrying single or multiple gene deletions. Half arrows are substrate-product relationships; solid connectors represent direct interactions between components, dashed connectors are inferred or indirect links. The references supporting these interactions are included in Supplemental Table 3.
