Figure 8.
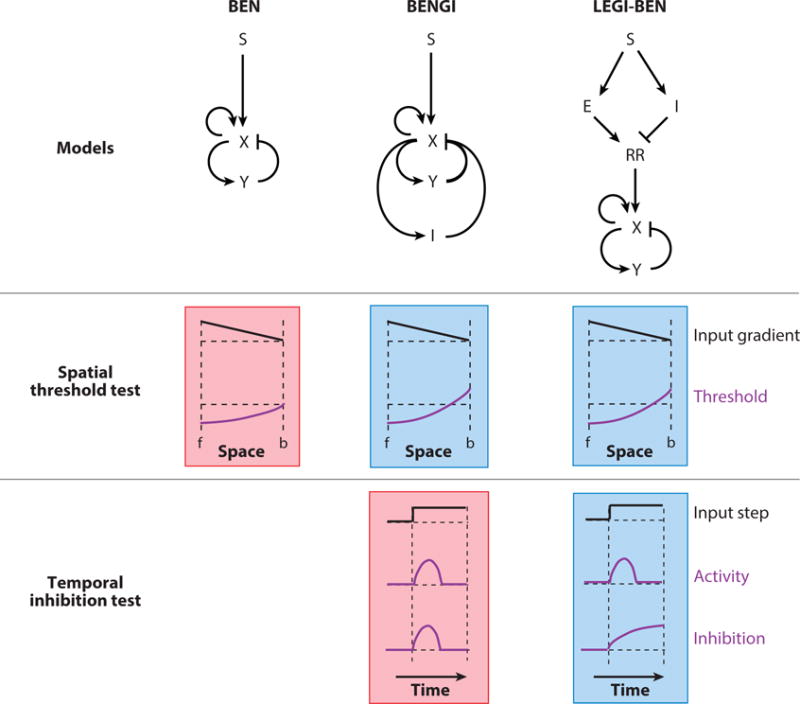
A cartoon of how LEGI-BEN emerged as a realistic model of gradient sensing. Three such proposed models (structures shown on top: S, external input; X and Y, activator and inhibitor of the excitable system; I, global inhibition; E; local excitation; RR, response regulator) are put through two specific tests. The outputs shown schematically represent the responses observed. The green and red shading represent passed and failed tests respectively. For the spatial threshold test, when an input gradient is applied (f: front, b: back of cell), all three models display a lowered threshold at the front. In the BENGI and LEGI-BEN models, which have global inhibitors, the threshold is raised at the back. This would result in better directed movement. To distinguish between these two, the cells were put through a temporal inhibition test, where the global inhibitor level is quantified for a homogenous input of chemoattractant. For the BENGI model, as the inhibition follows activator level, it dies down with time. However, in the LEGI-BEN scheme, the inhibitor is derived from the input itself causing the level of inhibition to rise with time as the step is sustained, matching real cell observations.
