Figure 1.
DsbA1 and DsbA2 Are Redundant and Required for Pleiotropic Phenotypes in S. marcescens
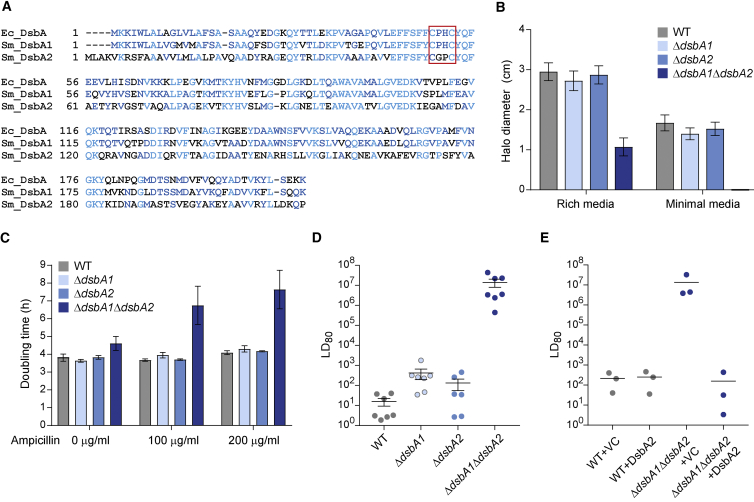
(A) Alignment of the amino acid sequences of DsbA1 (SMDB11_4101) and DsbA2 (SMDB11_0239) from S. marcescens Db10 with the single DsbA homolog from E. coli K12 (UniProt: P0AEG4). The conserved oxidoreductase catalytic motif (CXXC) is indicated by a red box.
(B) Swimming motility of wild-type (WT) or mutant (ΔdsbA1, ΔdsbA2, and ΔdsbA1ΔdsbA2) strains of S. marcescens Db10 in rich (LB) or minimal media, expressed as mean diameter of the swim halo ± SEM (n = 4).
(C) Ampicillin resistance of wild-type and mutant strains, as determined by doubling time during growth in minimal media with differing concentrations of ampicillin. Bars show mean ± SEM (n = 4).
(D) Virulence of wild-type S. marcescens Db10 and mutant strains in Galleria mellonella larvae. Data are expressed as lowest dose of bacteria required to kill 80% of the larvae (LD80) after 24 hr (with 20 larvae inoculated per group). Individual data points, overlaid with the mean ± SEM, are plotted for n = 7 independent experiments.
(E) Virulence phenotype of the wild-type and the ΔdsbA1ΔdsbA2 mutant carrying the vector control (+VC, pSUPROM) or a plasmid directing the expression of DsbA2 in trans (+DsbA2, pSC1507) measured as in (D), except that data are from n = 3 independent experiments.
See also Figure S1.