Abstract
The faecal flora is a common reservoir for urinary tract infection (UTI), and E. coli is frequently found in this reservoir without causing extraintestinal infection. We investigated these E. coli reservoirs by whole-genome sequencing a large collection of E. coli from healthy controls (faecal), who had never previously had UTI, and from UTI patients (faecal and urinary) sampled from the same geographical area. We compared MLST types, phylogenetic relationship, accessory genome content and FimH type between patient and control faecal isolates as well as between UTI and faecal-only isolates, respectively.
Comparison of the accessory genome of UTI isolates to faecal isolates revealed 35 gene families which were significantly more prevalent in the UTI isolates compared to the faecal isolates, although none of these were unique to one of the two groups. Of these 35, 22 belonged to a genomic island and three putatively belonged to a type VI secretion system (T6SS). MLST types and SNP phylogeny indicated no clustering of the UTI or faecal E. coli from patients distinct from the control faecal isolates, although there was an overrepresentation of UTI isolates belonging to clonal lineages CC73 and CC12.
One combination of mutations in FimH, N70S/S78N, was significantly associated to UTI, while phylogenetic analysis of FimH and fimH identified no signs of distinct adaptation of UTI isolates compared to faecal-only isolates not causing UTI
In summary, the results showed that (i) healthy women who had never previously had UTI carried faecal E. coli which were overall closely related to UTI and faecal isolates from UTI patients; (ii) UTI isolates do not cluster separately from faecal-only isolates based on SNP analysis; and (iii) 22 gene families of a genomic island, putative T6SS proteins as well as specific metabolism and virulence associated proteins were significantly more common in UTI isolates compared to faecal-only isolates and (iv) evolution of fimH for these isolates was not linked to the clinical source of the isolates, a part from the mutation combination N70S/S78N, which was correlated to UTI isolates of phylogroup B2. Combined, these findings illustrate that faecal and UTI isolates, as well as faecalonly and faecal-UTI isolates, are closely related and can only be distinguished, if at all, by their accessory genome.
Keywords: WGS, NGS, next generation sequencing, urinary tract infection, faecal flora, microbiota, gut, environment, virulence, fimbria, genomes, polymorphism, SNPs, mutations, phylogeny, evolution, adaption
Introduction
Escherichia coli is the most common cause of urinary tract infection (UTI) with the faecal flora of the patient as the primary source of the infecting isolate (Moreno et al., 2008; Nielsen et al., 2014; Yamamoto et al., 1997; Zhang et al., 2002). The faecal flora of healthy individuals and in UTI patients have been found to vary considerably. Some studies have described large proportions of phylogroup B2 and D isolates among UTI patients, different from healthy individuals who primarily carry A and B1 isolates (Duriez et al., 2001; Moreno et al., 2009). Others have described similar proportions of B2 and D isolates and virulence factors in healthy controls and UTI patients (Nielsen et al., 2014; Yamamoto et al., 1997; Zhang et al., 2002). The discrepancies illustrate great geographical variation in the prevalence of individual phylotypes (Bailey et al., 2010), but also possibly great differences in uropathogenicity across phylotypes, although phylotype B2 and D isolates belonging to clonal complexes CC69 (Tartof et al., 2005), CC73 (Johnson et al., 2006), CC127 (Johnson et al., 2006) and CC131 (Karfunkel et al., 2013) as determined by multi-locus sequence typing (MLST), constitute the majority of uropathogenic E. coli (UPEC). Several UPEC virulence factors are known, including type 1 fimbria, S fimbria, antigen 43 as well as iron scavengers. These virulence traits are also found in faecal isolates, which do not cause infection, and can therefore not be uniquely associated to pathogenicity (Bielaszewska et al., 2007; Dobrindt, 2005). It has, so far, not been possible to identify a common set of virulence factors that is unique to UTI (Ejrnæs et al., 2011; Mabbett et al., 2009; Norinder et al., 2011).
Type 1 fimbria are present in 80% of uropathogenic E. coli (UPEC) (Schembri et al., 2001). The tip of the fimbria, FimH, is produced as a precursor with an N-terminal signaling peptide of 21 amino acids, and the mature FimH (279 aa) protein consists of two domains: An N-terminal lectin domain (22–171 aa) as well as a C-terminal pilin domain (180–300 aa) connected by a 8 aa linker (Schwartz et al., 2013). Binding properties of FimH have been shown to be radically changed by mutations throughout the protein (Dreux et al., 2013; Hommais et al., 2003; Sokurenko et al., 2004, 1998, 1997), and data indicate that specific mutations in FimH are patho-adaptive (Schwartz et al., 2013). Adaptation of E. coli to the urinary environment is evident from several studies (Chen et al., 2009; Hommais et al., 2003; Schwartz et al., 2013; Sokurenko et al., 2004) and it has been proposed that this adaptation should be considered a dead-end, where the bacteria cannot successfully recolonize the original environment, due to adaptive changes making them unfit in e.g. the gut (Chattopadhyay et al., 2007; Sokurenko et al., 2004). In contrast, Chen et al. (Chen et al., 2013) identified the same E. coli from successive UTI episodes in both urine and faecal isolates, indicating that UPEC indeed could successfully colonize both bladder and gut. Additionally, we have recently shown that UPEC isolates show very few signs of adaptation compared to the faecal counterparts (Nielsen et al., 2016). This warrants an examination of genetic relationship between pathogenic and non-pathogenic isolates in more detail than phylogrouping and virulence typing can accomplish.
In this study a large collection of E. coli from healthy controls (faecal isolates), who have never had UTI, and UTI patients (faecal and urinary isolates) sampled from the same geographical area were whole-genome sequenced. Using phylogenetic analyses and detailed investigations of genetic content, we aimed at (i) characterizing whether UTI and faecal isolates can be distinguished at the genomic level and (ii) comparing the faecal E. coli of UTI patients with those of controls in order to describe whether the faecal E. coli of patients could predict UTI.
Methods
Study Participants
Participants and samples in this study have been described in detail (Nielsen et al., 2014a). Briefly, we recruited otherwise healthy pre-menopausal women with or without UTI from their own general practitioner in Zealand, Denmark. All participants delivered a urine sample and a faecal swab on the day of inclusion. Inclusion criteria for patients were UTI symptoms, ≥104 E. coli CFU/mL and leukocytes in their urine sample. Inclusion criteria for controls: Negative urine sample with respect to leukocytes and bacteria, and with no previous UTI infection. Exclusion criteria for both patients and controls were diabetes, pregnancy and post-menopause as well as women with recent urogenital surgery. A total of 47 UTI patients and 50 controls were included in the study (Nielsen et al., 2014a).
Bacterial Isolates
From each rectal swab, up to 20 E. coli isolates (if available) were picked and typed by RAPD PCR for identification of unique clones (Nielsen et al. 2014b). Picking 20 E. coli colonies from each sample will with estimated 88% certainty reveal all E. coli clones with as low as 10% prevalence in the sample (Lautenbach et al., 2008). The collection of bacterial isolates contained 196 isolates: 48 UTI isolates from 47 patients (one patient had two E. coli types in her urine) as well as 148 faecal isolates from patients (n=81) and controls (n=67) classified as major (>50%), minor (<10%) and intermediate (10 ≤ 50%) clones as described elsewhere (Nielsen et al., 2014a). Faecal isolates of patients were further classified as 41 faecal-only (faecal isolates of patients different from the infecting UTI clone) and 40 faecal-UTI isolates (faecal-isolates similar to a urinary isolate of the same individual). All isolates were phylo-grouped into phylogroups as described by Clermont et al. 2013 (Clermont et al., 2013) by BLAST in silico (Moriel et al., 2016). For C- and E-specific primers, only 100% primer matches were considered hits.
Whole-genome Sequencing
DNA was purified using DNeasy Blood and Tissue kit (Qiagen). The isolates were subjected to whole-genome sequencing on a HiSeq 2000 Instrument (Illumina). All isolates were sequenced using 180-bp paired-end fragment libraries and 3-kb paired-end jumping libraries (mate pair) (Grad et al., 2012). Exception to this was forty-two faecal isolates predicted identical to the infecting UTI isolate based on RAPD typing and phylogroups which were sequenced from 180-bp fragment libraries only (Nielsen et al., 2016).
Assembly and Annotation
Assemblies were generated using ALLPATHS-LG (Butler et al., 2008; Gnerre et al., 2011), and gene contents were grouped into orthologous clusters to calculate number of clustered genes across the complete set of genomes using reciprocal best hit BLAST. Core genes were defined as present with at least one copy in the genomes of all included isolates, accessory genes were defined as absent in the genome of at least one isolate. The assembled genomes consisted of 2–189 scaffolds (average 28). Open reading frames were identified in all genomes using Prodigal, as described by Grad et al. (Grad et al., 2012). Annotations were based on top BLAST hits against Swiss-Prot database (≥70% identity and ≥70% query coverage), TIGRfam and Pfam.
Phylogenetics
A maximum likelihood phylogeny was built using identified single nucleotide polymorphisms (SNPs) were determined in the core set. This was performed for the 48 UTI isolates and the 108 unique faecal isolates (with the faecal-UTI isolates excluded) (N=156). SNPs were identified by first aligning reads to the reference E. coli UTI89 (GenBank accession: NC_007946) using BWA (Li and Durbin, 2009) version 0.7.4 and variants were called using GATK (Mckenna et al., 2010) version 2.5.2. Any site with ambiguous or missing call or depth less than 10 in any sample was removed. Areas of likely recombination were removed by identifying consecutive variant patterns that were longer than would be expected at random, at a 5% cutoff level using Bonferroni correction for multiple testing. Phylogenetic reconstruction was performed using dnaml from the PHYLIP package (Felsenstein, 1989) with default parameters.
Accessory Genome Analyses
The accessory genome was compared by three different analyses: (i) total patient faecal isolates (faecal-only and faecal-UTI isolates, N=81) vs. control faecal isolates (N=67), (ii) faecal-only isolates of patients (N=41) vs. control faecal isolates (N=67), and (iii) UTI isolates (N=48) vs. faecal-only isolates (from patients and controls combined, N=108). Protein sequences were extracted from the genomes using Prodigal and searched against the Pfam-A database (Punta et al., 2012; Sonnhammer et al., 1997) using HMMER3 (Eddy, 2011). The protein sequences were grouped by absence or presence of Pfam protein domains. Within each group, all sequences were compared pairwise using BLAST version 2.2.26 (Boratyn et al., 2012) without low-complexity filtering and using an E-value cutoff of 0.05. Sequences of each group were subsequently clustered based on BLAST matches using the Markov Cluster (MCL) algorithm (Dongen, 2008; Enright et al., 2002) with a set I value of 5. Stable clusters were considered gene families. Significance was determined by two-tailed Fisher’s exact test for each gene family adjusted for multiple testing using Benjamini and Hochbergs false discovery rate as implemented in the R Stats package.
In silico MLST Typing
MLST types were assigned in silico by the scheme of Wirth et al. (Wirth et al., 2006). MLST alleles, which were downloaded from www.mlst.net, were blasted against the genome assembly, and the allele with the closest match (usually 100% identity) for each MLST locus was reported. If all seven MLST loci belonged to a known MLST type, then this type was reported. MLST types were assigned to clonal complexes (CC) using eBURST V3 (eburst.mlst.net).
fimH Characterization
All complete fimH were identified in the assembled genomes of the faecal and UTI isolates and were translated and aligned using CLC Main Workbench version 6.5.2 (CLC bio) for investigation of the sequence diversity. Any variation observed in four or more isolates across the protein sequences was compared to the clonal status of the isolate. A maximum-likelihood phylogeny on the amino acid sequences of all complete FimH was created using CLC Main Workbench.
A maximum likelihood phylogenetic tree comparing the nucleotide sequences of all complete fimH was created using K80 nucleotide substitution model and 100 bootstrap using CLC Main Workbench 6.5.2 (CLC bio). Proportions of FimH mutations and clonal complexes were compared applying two-tailed Fischer’s exact test. Correlation of FimH mutations to source of the isolate was evaluated by the χ2 test for independence followed by relevant post-test by two-tailed Fisher’s exact test (P<0.016, Bonferroni corrected).
Results
The Faecal E. coli Population
Analyses of the accessory genome of patient faecal isolates and control faecal isolates showed no significant differences (data not shown). We did not observe any differences of the MLST distribution between faecal isolates from patients and controls (Table 2) and a core SNP analysis showed no distinct clustering of faecal isolates of patients compared to faecal isolates of controls (Figure 1).
Table 2.
MLST types assigned to clonal complexes of faecal isolates of control, patients and UTI isolates.
| Clonal complex |
Control N=67, n (%) |
Patient§ N=81, n (%) |
Non-UTI‡ N=108, n (%) |
UTI N=48, n (%) |
|---|---|---|---|---|
| CC69 | 7 (11) | 9 (11) | 8 (7) | 8 (17) |
| CC10 | 12 (18) | 11 (14) | 20 (19) | 3 (6)† |
| CC12 | 0 (0) | 2 (2) | 0 (0) | 3 (6)* |
| CC127 | 1 (2) | 1 (1) | 1 (1) | 1 (2) |
| CC131 | 2 (3) | 4 (5) | 5 (5) | 1 (2) |
| CC14 | 1 (2) | 3 (4) | 1 (1) | 3 (6) |
| CC141 | 4 (6) | 4 (5) | 5 (5) | 4 (8) |
| CC357 | 4 (6) | 1 (1) | 4 (4) | 1 (2) |
| CC394 | 0 (0) | 2 (2) | 1 (1) | 1 (2) |
| CC399 | 1 (2) | 2 (2) | 3 (3) | 0 (0) |
| CC59 | 5 (7) | 2 (2) | 7 (6) | 0 (0) |
| CC73 | 4 (6) | 9 (11) | 6 (5) | 10 (21)* |
| CC95 | 9 (13) | 5 (6) | 12 (11) | 3 (6) |
| none | 5 (7) | 9 (11) | 11 (10) | 3 (6) |
| singleton | 4 (6) | 2 (2) | 6 (6) | 1 (2) |
| ≤2 of same CC | 8 (12) | 15 (19) | 18 (17) | 6 (13) |
Patient faecal-only and faecal-UTI clones
Includes faecal clones of controls as well as faecal-only clones of patients
significantly associated to UTI (P<0.05)
Borderline significant, P=0.05
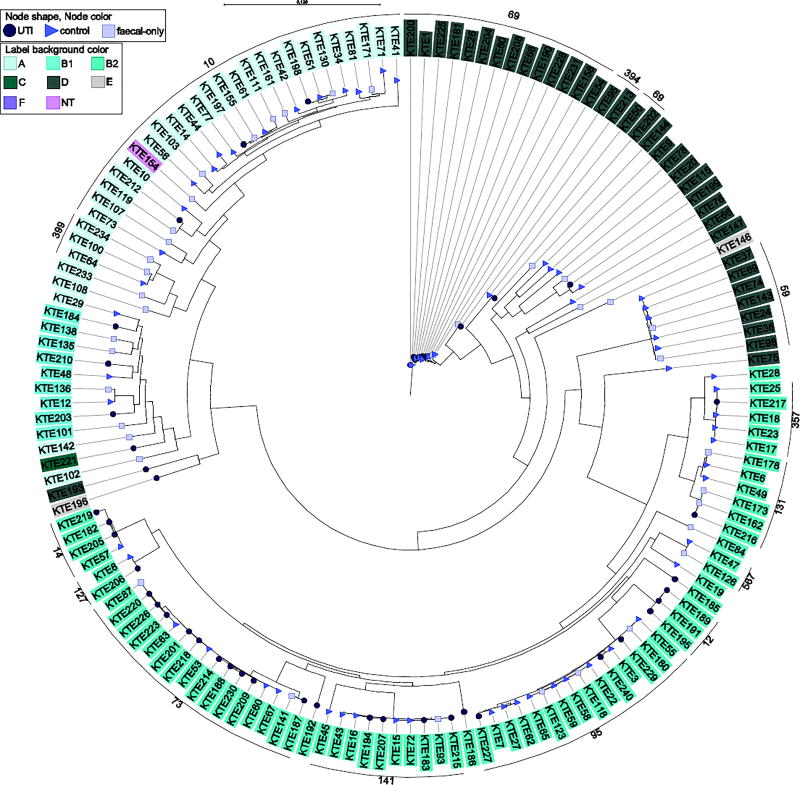
Figure 1.
Phylogenetic reconstruction of 156 urinary and fecal E. coli isolates. Clustering of major clonal complexes has been assigned (black line along periphery). Phylogroups and source of the isolates are indicated with background colour and bullet shape and colour, respectively
The phylogeny showed general clustering according to phylogroups and even better clustering of MLST clonal complexes (Figure 1). A group of phylogroup F isolates were very different from the other E. coli phylotypes, indicating the great degree of diversity of E. coli (Supplementary figure S1).
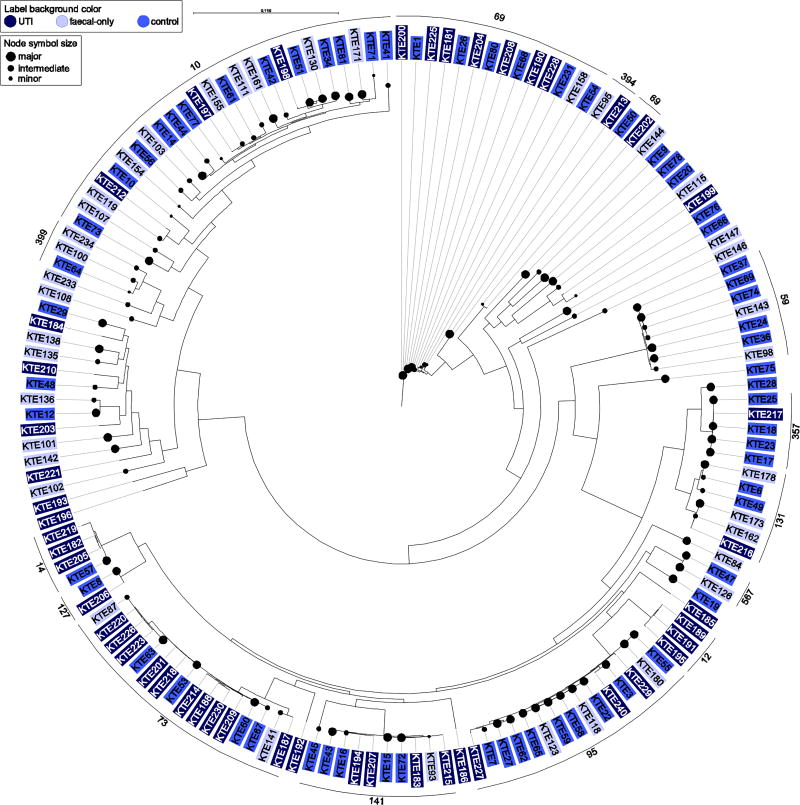
We investigated distribution of major (>50%), minor (<10%) and intermediate (10 ≤ 50%) faecal clones in the phylogeny. With respect to faecal dominance, ST95 was significantly associated to major and intermediate faecal clones (Fischer’s exact test, P=0.001), whereas ST399 contained minor and intermediate isolates only (n=3, not significant). ST357 (n=4) and ST567 (n=2) contained major faecal clones, and ST131 contained isolates of both major (n=2) and minor clone status (n=3, not significant) (Figure 2).
Figure 2.
Phylogenetic reconstruction of 156 urinary and fecal E. coli isolates. Faecal dominance is indicated by bullet size. Clustering of major clonal complexes has been assigned (black line along periphery).
Faecal isolates compared to UTI isolates
In order to identify possible genetic differences between isolates only found in faeces and isolates which caused UTI, we compared the accessory genome of these two groups. The analyses revealed that 38 gene families were significantly more common either in the UTI isolates (OR<1, n=35) or faecal isolates (OR>1, n=3) (Table 1, Figure 3).
Table 1.
Gene products with significant differences in proportions between faecal-only and UTI isolates
| # | Function | Locus tag1 | Nt position1 | q- value2 |
OR3 | Faecal N=108 n (prop4) |
UTI N=48 n (prop4) |
|---|---|---|---|---|---|---|---|
| 1* | 3-oxoacyl-reductase (Fatty acid and phosphatidic acid biosynth) | C1187 | 1,145,684–1,144,953 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 2* | O-methyltransferase | C1203 | 1,157,236–1,158,294 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 3* | FabA-like domain | C1187 | 1,145,684–1,146,148 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 4* | thioesterase | C1193 | 1,150,789–1,151,208 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 5* | outer membrane lipoprotein carrier protein LolA | C1192 | 1,150,184–1,150,789 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 6 | hypothetical protein | C1202 | 1,156,473–1,157,219 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 7 | hypothetical protein | C1198 | 1,154,574–1,155,158 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 8 | hypothetical protein | C1204 | 1,158,359–1,158,748 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 9* | beta-ketoacyl-ACP synthase | C1186 | 1,144,956–1,143,727 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 10* | phosphopantetheine attachment site (Fatty acid and phosphatidic acid biosynth) | C1200 | 1,154,423–1,155,680 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 11* | phosphopantetheine attachment site (Fatty acid and phosphatidic acid biosynth) | C1199 | 1,155,160–1,155,411 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 12 | hypothetical protein (MMPL family) | C1191 | 1,147,897–1,150,215 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 13* | AMP-binding enzyme (Fatty acid and phosphatidic acid biosynth) | C1197 | 1,153,219–1,154,577 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 14 | domain of unknown function | C1190 | 1,147,316–1,147,900 | 0.035 | 0.23 | 20 (0.19) | 24 (0.50) |
| 15 | hypothetical protein | C1209 | 1,166,202–1,166,597 | 0.035 | 0.16 | 9 (0.08) | 18 (0.38) |
| 16 | hypothetical protein | CFT073 | 1,164,857–1,165,177 | 0.035 | 0.16 | 9 (0.08) | 18 (0.38) |
| 17 | domain of unknown function | CFT073 | 1,165,927–1,166,106 | 0.035 | 0.16 | 9 (0.08) | 18 (0.38) |
| 18 | hypothetical protein | C1208 | 1,165,325–1,165,702 | 0.035 | 0.18 | 12 (0.11) | 20 (0.42) |
| 19 | microcin M activity protein (CAAX amino terminal Protease) | C1233 | 1,183,572–1,184,258 | 0.035 | 0.20 | 14 (0.13) | 21 (0.44) |
| 20 | partial putative transposase | C1219 | 1,171,141–1,171,377 | 0.035 | 0.16 | 7 (0.07) | 15 (0.31) |
| 21 | domain of unknown function | C1190 | 1,147,316–1,147,900 | 0.035 | 0.16 | 7 (0.07) | 15 (0.31) |
| 22 | putative type VI secretion system protein 1, T6SS2 gene locus | ECUMN_0233 | 267,016–268,026 | 0.035 | 0.17 | 13 (0.12) | 22 (0.46) |
| 23 | putative type VI secretion system protein 2, T6SS2 gene locus | ECUMN_0232 | 265,378–266,949 | 0.036 | 0.21 | 14 (0.13) | 20 (0.42) |
| 24 | putative type VI secretion system protein 3, T6SS2 gene locus | ECUMN_0231 | 264,515–265,356 | 0.035 | 0.18 | 12 (0.11) | 20 (0.42) |
| 25 | Hypothetical protein | C1209 | 1,166,202–1,166,597 | 0.035 | 0.16 | 9 (0.08) | 18 (0.38) |
| 26 | HNH nuclease + S-type Pyocin | UTI89_C4900 | 4,800,111–4,800,587 | 0.035 | 0.17 | 9 (0.08) | 17 (0.35) |
| 27 | putative DNA-binding transcriptional regulator | APECO78_06720 | 1,380,572–1,380,949 | 0.035 | 0.21 | 20 (0.19) | 25 (0.52) |
| 28 | transposase + Putative transposase DNA-binding domain + HTH domain | ECOLIN_04005 | 834,611–835,741 | 0.035 | 0.21 | 17 (0.16) | 23 (0.48) |
| 29* | HisKA + ATPase domain | C0780 | 762,465–763,943 | 0.036 | 0.20 | 12 (0.11) | 19 (0.40) |
| 30* | shiA homolog: polysaccharide export protein VexE | C4492 | 4,276,297–4,277,340 | 0.035 | 0.22 | 49 (0.46) | 38 (0.79) |
| 31 | prophage CP4-57 regulatory protein (AlpA) | UTI89_C4952 | 4,858,346–4,858,579 | 0.035 | 0.21 | 38 (0.36) | 35 (0.73) |
| 32 | putative transposase ORF2, IS66 Family | C0257 | 250,214–250,564 | 0.036 | 0.24 | 42 (0.39) | 35 (0.73) |
| 33* | phosphotransferase system: glucose-specific EIIB+EIIC component | C4758 | 4,522,574–4,524,082 | 0.036 | 0.24 | 47 (0.44) | 37 (0.77) |
| 34* | molo-1 founding proteins of phosphatase | C3340 | 3,180,005–3,181,159 | 0.035 | 0.22 | 35 (0.33) | 33 (0.69) |
| 35 | toxin SymE (Toxin/antitoxin system) | C5422 | 5,164,819–5,165,160 | 0.035 | 0.14 | 59 (0.55) | 43 (0.90) |
| 36 | domain of unknown function in dinQ-agrB toxin/antitoxin system | APEC078 | 4,404,064–4,404,237 | 0.036 | 4.50 | 89 (0.83) | 25 (0.52) |
| 37* | 4-oxalocrotonate tautomerase | APEC078_11025 | 2,270,508–2,270,782 | 0.035 | 4.77 | 63 (0.59) | 11 (0.23) |
| 38 | bacteriophage Rz lysis protein | APEC078_03145 | 645,810–646,271 | 0.035 | 6.92 | 48 (0.45) | 5 (0.10) |
Locus Tag and nt position based on CFT073 (GenBank accession AE14075) with the following exceptions: 22-24; UMN026 (CU928163), 26+31; UTI89 (CP000243), 28; Nissle 1917 (CP007799), 27 + 36-38; APEC078 (CP004009)
q-value: adjusted p-value after adjusting for multiple testing.
OR: Odds ratio, OR<1: overrepresented in UTI isolates, OR>1: overrepresented in faecal isolates.
Prop: proportion of UTI and faecal-only isolates respectively.
Proteins involved in metabolism.
Italic and grey: Proteins encoded on genomic island I of E. coli Nissle 1917.
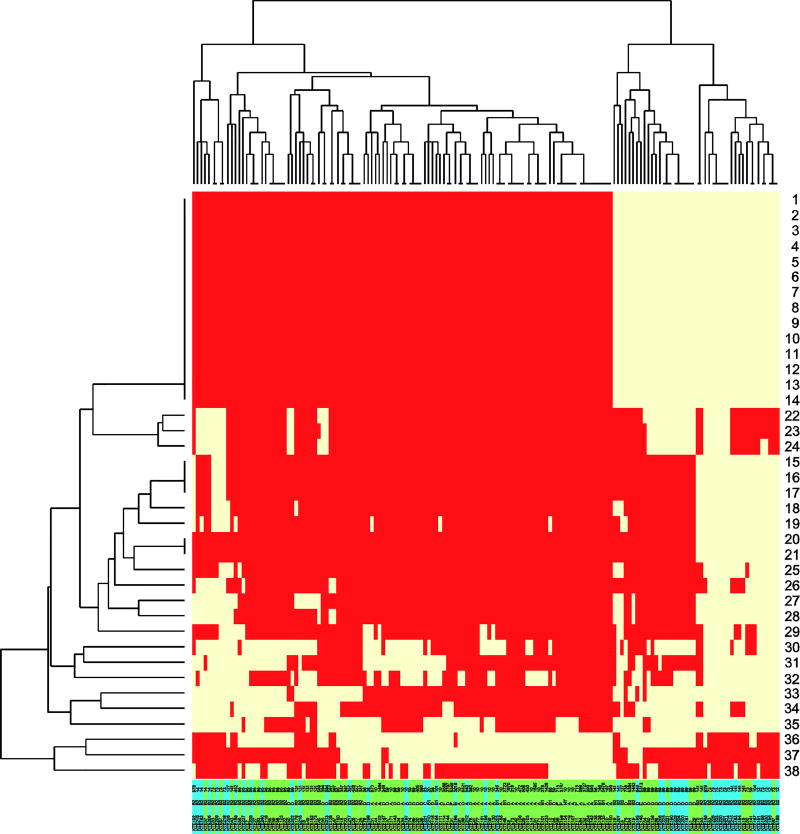
Figure 3.
Heatmap of proteins (1–38, details in Table 1) significantly overrepresented in UTI or faecal isolates. Yellow: Presence, red: Absence. Colouring under branches: green: faecal-only isolates (from controls and patient faecal-only isolates), blue: UTI isolates. Protein family 1–21 and 25 are encoded on genomic Island I of E. coli Nissle 1917. Protein family 22–24 are encoded in the same gene locus as T6SS.
Of the 35 gene families which were overrepresented in the UTI isolates, 22 (marked in italic and grey in Table 1) constitute a part of genomic island I (GEI I), a genomic island originally identified in E. coli Nissle 1917; an apathogenic E. coli isolate. The genes encoding the significant proteins were distributed in the first 45kb of the island (Figure 6) and presence of the 22 genes was linked as evident in the heatmap (Figure 3). In both faecal and UTI isolates we observed variants of this island of varying lengths, including all or some of the genes significantly associated to UTI (Table 1, Figure 3). These 45kb were also present in other UTI-related isolates, including CFT073 (AE014075), ABU 83972 (CP001671), but also ATCC 25922 (CP009072).
Figure 6.
Genomic Island I of E. coli Nissle 1917, modified from GenBank: AJ586887 with annotations identified significant in this study. Top: Complete GEI I (97552 bp), dark blue indicate the gene products associated to UTI. Bottom: 1–46000bp of GEI with annotations. HP: hypothetical protein. 37,000–46,000bp: Microcin genes.
Three of the 35 UTI associated proteins were gene families encoding proteins putatively belonging to type VI secretion system (T6SS) and carriage of these three gene families were linked due to localisation in the same gene locus (Table 1, Figure 5). Thirty-two isolates carried all three genes, of which 22 isolates carried the complete T6SS as well as the three putative T6SS genes and 10 isolates carried the three putative genes only. The GEI I elements and the three putative T6SS proteins significantly correlated to UTI were found in B2 and D isolates only (Figure 3). The remaining proteins over-represented in the UTI group included both virulence and metabolic proteins (Table 1).
Figure 5.
Overview of T6SS gene cluster spanning from Hcp protein to Rhs protein (Genbank: JN837480). Notably, the Rhs protein overlaps with the first of the putative T6SS proteins.
The comparison of UTI and faecal isolates also identified 4 suspicious gene families (domains of unknown function (DUF) in table 1). These predicted gene family products contained only domains of unknown function, and the genes were placed both inside and outside known annotations in well annotated genomes: In E. coli Nissle DUFs were outside predicted open reading frames, however in CFT073 two of the four were predicted as hypothetical proteins (#14 and 21, Table 1). The open reading frames received mid to low scores in Prodigal, and hence, the significance of these could indicate presence of genomic islands, rather than actually expressed proteins, in particular for the ones placed outside open reading frames.
The comparison of accessory genome between UTI and faecal isolates also revealed three protein families, which were overrepresented in the faecal isolates compared to the UTI isolates; a toxin/antitoxin system, a tautomerase and a bacteriophage lysis protein (Table 1, Figure 3). Contrary, the analyses did not identify any of the classical UPEC virulence factors e.g. fimbria and siderophores as significantly overrepresented in UTI isolates compared to the faecal-only isolates.
CC73 and CC12 were significantly more common among UTI isolates when compared to faecal-only isolates (P<0.05) (Table 2). Additionally, CC10 was more common in faeces, however, only borderline significant (P=0.05). The phylogenetic tree did not show any distinct clustering of UTI isolates (Figure 1); however, CC59 (n=7) was unique to faecal isolates and these clustered separately in the phylogenetic tree (Figure 1).
The Importance of fimH
Forty-seven (98%) UTI isolates, 62 (92.5%) faecal isolates from controls and 37 (90%) patient faecal-only isolates were found to carry fimH. The proportion of isolates with fimH was not significantly different between UTI isolates and faecal isolates which did not cause infection (P=0.27).
We identified sites with mutations in FimH of UTI and faecal isolates and identified the mutation N70S significantly associated to UTI (Table 3). Additionally, N70S was linked to S78N for the UTI isolates; 18 UTI isolates (38%) and 18 faecal isolates (18%) (P=0.01).
Table 3.
FimH mutations in faecal and urinary isolates.
| Position* | Control N=62, n (%) |
Faecal-only N=37, n (%) |
Faecal total N=99, n (%) |
UTI N=47, n (%) |
|---|---|---|---|---|
| T6N (SP) | 7 (11) | 3 (8) | 10 (10) | 5 (11) |
| T6Y (SP) | 1 (2) | 0 (0) | 1 (1) | 0 (0) |
| A27V | 11 (18) | 5 (14) | 16 (16) | 14 (30) |
| A27T | 0 (0) | 1 (3) | 1 (1) | 0 (0) |
| G66S | 1 (2) | 1 (3) | 2 (2) | 1 (2) |
| G66R | 0 (0) | 1 (3) | 1 (1) | 1 (2) |
| N70S | 13 (21) | 5 (14) | 18 (18) | 18 (38) † |
| S78N | 22 (35) | 9 (24) | 31 (31) | 19 (40) |
| N70S/S78N | 13 (21) | 5 (14) | 18 (18) | 18 (38)† |
| A119V | 5 (8) | 3 (8) | 8 (8) | 5 (11) |
| S139G | 2 (3) | 2 (5) | 4 (4) | 0 (0) |
| V163A | 1 (2) | 0 (0) | 1 (1) | 4 (9) |
| R166S | 1 (2) | 1 (3) | 2 (2) | 0 (0) |
| R166H | 5 (8) | 2 (5) | 7 (7) | 4 (9) |
| G205D | 2 (3) | 2 (5) | 4 (4) | 0 (0) |
| S236N | 3 (5) | 3 (8) | 6 (6) | 0 (0) |
| A242V | 2 (3) | 2 (5) | 4 (4) | 0 (0) |
| Q269K | 8 (13) | 5 (14) | 13 (13) | 1 (2) |
| G273A | 2 (3) | 3 (8) | 5 (5) | 4 (9) |
Positions are given as positions in fimH excluding the signaling peptide.
P=0.01
SP: signaling peptide.
The wild-type allele was defined as the most frequently isolated allele.
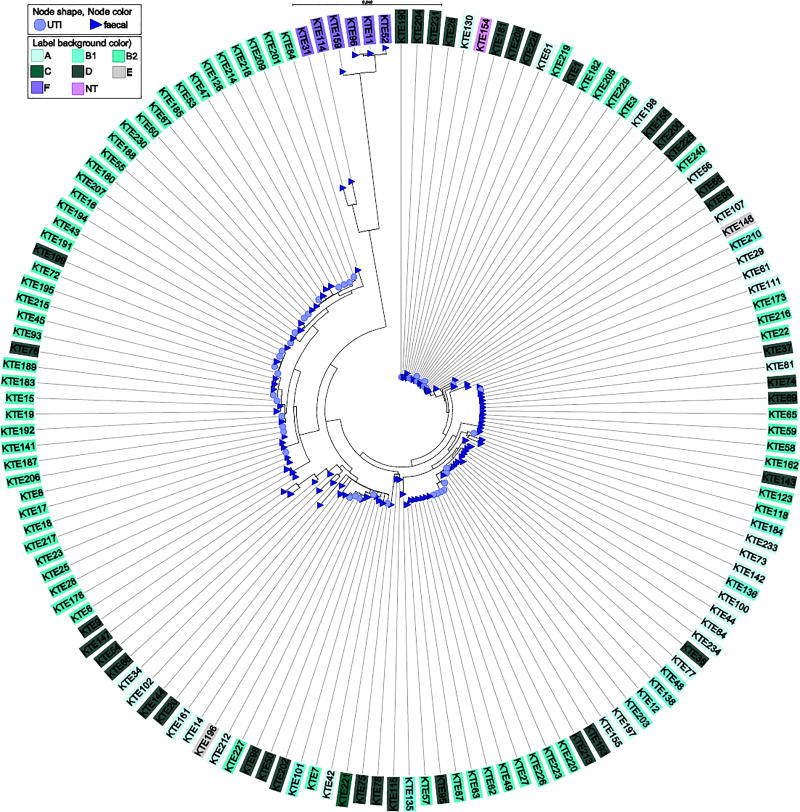
A phylogeny based on the nucleotide sequences of fimH (Figure 4) illustrated that (i) UTI and faecal isolates, which did not cause infection, did not cluster separately, (ii) some parts of the tree cluster with respect to phylogroups, e.g. the large proportion of B2 isolates (iii) variation at the nucleotide level was much greater than that observed at protein level, specifically with respect to the diverse NT cluster (Supplementary Figure S2).
Figure 4.
Maximum-likelihood fimH phylogeny. Faecal isolates constitute control isolates and faecal-only isolates of patients.
Discussion
In this study, we sought to compare the complete genome of E. coli with the aim of identifying both clonal clustering as well as specific genetic factors, which are important for UTI. With high confidence, we analysed the majority of all faecal clones from each rectal swab, hence, the current study yields a very thorough analysis of the clone prevalence in faecal isolates of UTI patients and healthy controls.
The definition of a virulence factor is in some cases arbitrary and the success of one isolate is not necessarily dependent on direct virulence ability, rather also includes metabolism and fitness in the local environment (Chen et al., 2013; Grozdanov et al., 2004). This study is, to our knowledge, the first to compare the isolates from the faecal flora of healthy controls, who have not had UTI previously to those of patients with UTI, based on a large collection of faecal and UTI E. coli isolates sampled from the same geographical area.
Can the faecal flora predict UTI?
MLST, SNP-based phylogeny and accessory genome analyses showed that healthy women who had never had UTI carried very similar faecal E. coli to UTI patients without this causing infection. This demonstrates that faecal isolates in UTI patients were not of a certain ancestral type compared to commensal isolates which did not cause infection in healthy controls.
The present data indicate that faecal dominance (major, minor and intermediate clones) could overall not be identified based on molecular typing or phylogenetically, however, for a few MLST types there was a trend of major/minor clones clustering. The faecal dominance is likely to depend on host immune status, faecal flora composition and possibly the gene repertoire of the bacteria.
Can UTI isolates be distinguished from faecal-only isolates?
We identified 35 proteins which were overrepresented in the UTI isolates compared to the faecal isolates that did not cause infection, although UTI isolates could not be distinguished from faecal isolates based on SNP analysis.
The accessory genome analysis identified 19 proteins and three DUFs which were correlated to each other and known to be situated on GEI I of E. coli Nissle 1917 strain, indicating that this partial island contributes to fitness or virulence of the UTI isolates. This island has been described to be common in ExPEC, but less frequent in non-pathogenic faecal isolates of healthy humans (Grozdanov et al., 2004), which is in line with our findings. We identified Microcin M activity protein (McmM) to be significantly correlated to UTI. Microcin expression is encoded by a part of GEI I of E. coli Nissle 1917 and has been described as important for intestinal binding and hence, competition with other bacteria (Grozdanov et al., 2004). This could indicate that strong intestinal colonisation is an important utility of UTI isolates, however, this warrants further future investigations.
T6SS injects effector proteins into the cytosol of eukaryotic and bacterial cells in a bacteriophage-like manner (Hood et al., 2011). The system has previously been correlated to pathogenesis of avian pathogenic E. coli (APEC), which are closely related to ExPEC and represents a potential zoonotic risk to humans and a virulence reservoir for ExPEC (de Pace et al., 2011, 2010). Isolates with a non-functional T6SS have been shown to have decreased adherence, due to less expression of type 1 fimbriae, and lower invasion of epithelial cells, decreased intra-macrophage survival as well as lower biofilm formation (de Pace et al., 2011, 2010). These utilities also contribute to UTI virulence indicating that T6SS could play a role in UTI development.
Three putative proteins of T6SS were identified in the accessory genome faecal-only comparison with UTI isolates, indicating that this could be a new and previously unidentified predictor for UTI. The ORFs of the putative T6SS gene are adjacent to efvW (Rhs protein) of T6SS and overlap in nucleotide sequence (Figure 5). A protein BLAST search yields results with annotations of the three proteins as hypothetical proteins and ‘putative type VI secretion system protein’ in Escherichia coli 042 (CBG33062.1). The three putative T6SS elements were found in phylogroup B2 and D only (22–24 in Figure 3), the phylogroups which represent the majority of UTI isolates. This indicates that the putative T6SS genes are ancestrally linked to these phylogroups. The BLAST result and position of the genes indicate that the three genes could be correlated to T6SS. On the other hand, Rhs proteins are sites of insertions (Koskiniemi et al., 2013), so the role in UTI virulence of the three putative T6SS genes should be investigated further in order to determine their function.
Specific metabolism proteins were overrepresented in the UTI isolates compared to the faecal isolates (marked with asterisk in Table 1) (nine of the genes from GEI I as well as five of the eleven protein families which were not associated to neither GEI I or the T6SS locus). This indicates that specific factors of metabolism possibly aid to a higher UPEC fitness in the urinary tract compared to faecal isolates, which did not cause infection, underlining the importance of investigating broadly when studying UTI correlated factors and not merely investigating known and UTI relevant virulence genes.
Chen et al. (Chen et al., 2013) studied faecal and urinary E. coli from four patients and concluded that UPEC appear to be capable of existing in both the faecal and urinary environment without a fitness cost. Our results support and elaborate these results on a larger collection of clinical isolates. Additionally, the results are in line with our recent study of 42 paired faecal and UTI isolates from 42 patients from the same collection of isolates which showed minimal adaptation when comparing the UTI isolate and the faecal counterpart, indicating that the UTI isolate is fit in both the gut and urinary tract (Nielsen et al., 2016). A potential limitation to this interpretation on the bacterial adaption to the environment is the lack of information on intra-clonal diversity within either environment. However, we recently specifically investigated this variation (Nielsen et al., 2016) and found it to be minimal, and thus, was not further investigated here.
The results of this study question a distinction of commensal faecal isolates and ExPEC isolates as two separate E. coli types, as these share substantial parts of the genetic backbone and cannot be distinguished in a SNP phylogeny. There was, however, differences in the accessory genome i.e. overrepresentation of metabolism genes, genes of GEI I and putative T6SS genes, suggesting that UTI development depends on virulence, ability to bind to intestinal epithelial cells as well as fitness of the isolate.
The importance of FimH
fimH mutation T6N has previously been correlated to UTI isolates. In the current study, with a large collection of clinical isolates, we observed only few signs of patho-adaptive mutations in the UTI isolates and no correlation of T6N with UTI. Previous studies have suggested that the FimH mutations A48V, A62S and G66R affect the binding affinity of FimH and N70S in combination with S78N has been correlated to UTI isolates without effect on the binding affinity (Aprikian et al., 2007; Chen et al., 2009; Schwartz et al., 2013; Sokurenko et al., 1995; Weissman et al., 2007). We identified N70S/S78N in FimH significantly more often in UTI isolates compared to faecal isolates. This mutation was correlated to B2 (94%) and CC73 (44%) and clustered with the B2 isolates (Figure 1), but was identified in both faecal and urinary isolates. Our study of the core content and FimH of a large collection of faecal and urinary E. coli indicates that UTI isolates are likely to be well adapted to both the gut and urinary environment and that no significant adaptation occurred in the UTI isolates compared to the faecal isolates. Based on this, we consider the mutation combination N70S/S78N as stable in a successful UTI clone of phylogroup B2 and CC73 rather than a sign of pathogenic adaptation, which is supported by Chen et al. (Chen et al., 2009). Discrepancies between the present and previous studies could be due to different source populations or sampling strategies, but could also indicate that the faecal isolates of this collection of E. coli are capable of causing UTI.
It has been suggested that horizontal gene transfer is a common mediator of the variation found in fimH (Hommais et al., 2003). This is supported by the knowledge of the fim cluster being situated in a highly recombinogenic part of the E. coli genome (Touchon et al., 2009), into which pathogenicity islands often integrate (Weissman et al., 2012). The observation that phylotypes and FimH alleles are not clustered, leads us to conclude that the evolution of FimH in the individual isolates in this study was due to both adaptive mutations and horizontal gene transfer; a hypothesis supported by Hommais et al. (Hommais et al., 2003).
A recent study by Dreux et al. (Dreux et al., 2013) characterising adherent invasive E. coli in correlation to Crohn’s disease has identified presence of the same FimH mutations as found in this study, and identified that the clade with the mutation N70S/S78N was correlated to increased binding to T84 epithelial cells. This combination of mutations was in this study significantly more common in the UTI isolates compared to the faecal isolates, and indicates that fimH aids strong intestinal binding prior to UTI development as well as the well-characterised role during adherence to uroepithelial cells.
Concluding remarks
The key findings of this study were that UTI isolates do not cluster distinctly from faecal-only isolates in a phylogeny, indicating that UTI isolates did not evolve distinctly from commensal E. coli. UTI isolates more commonly contained genes associated to a genomic island, three putative genes of T6SS and specific metabolism genes. Faecal E. coli of patients and controls could not be distinguished in a phylogeny or in the accessory genome. Acquisition of a pathogenic E. coli is not the rate-limiting step of the UTI infection, but rather, a combination of factors, including ability to bind to intestinal epithelial cells, virulence, metabolism, fitness of the isolate and immune status of the host.
Supplementary Material
Acknowledgments
The authors would like to thank the technical staff and doctors at Haslev Lægecenter and Lægehuset Ellemarksvej for excellent technical assistance and a fruitful collaboration. This work was a part of Predicting Antibiotic Resistance (PAR), an EU FP7-Health-2009-Single-Stage project (grant 241476). Additionally, the work was supported by The Danish Council for Strategic Research (DanCARD project 09-067075/DSF). This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No.: HHSN272200900018C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aprikian P, Tchesnokova V, Kidd B, Yakovenko O, Yarov-Yarovoy V, Trinchina E, Vogel V, Thomas W, Sokurenko E. Interdomain interaction in the FimH adhesin of Escherichia coli regulates the affinity to mannose. J. Biol. Chem. 2007;282:23437–46. doi: 10.1074/jbc.M702037200. [DOI] [PubMed] [Google Scholar]
- Bailey JK, Pinyon JL, Anantham S, Hall RM. Distribution of human commensal Escherichia coli phylogenetic groups. J. Clin. Microbiol. 2010;48:3455–6. doi: 10.1128/JCM.00760-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M, Dobrindt U, Gärtner J, Gallitz I, Hacker J, Karch H, Müller D, Schubert S, Alexander Schmidt M, Sorsa LJ, Zdziarski J. Aspects of genome plasticity in pathogenic Escherichia coli. Int. J. Med. Microbiol. 2007;297:625–39. doi: 10.1016/j.ijmm.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Boratyn GM, Schäffer Aa, Agarwala R, Altschul SF, Lipman DJ, Madden TL. Domain enhanced lookup time accelerated BLAST. Biol. Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J, Maccallum I, Kleber M, Shlyakhter IA, Belmonte MK, Lander ES, Nusbaum C, Jaffe DB. ALLPATHS : De novo assembly of whole-genome shotgun microreads. 2008:810–820. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Feldgarden M, Weissman SJ, Dykhuizen DE, van Belle G, Sokurenko EV. Haplotype diversity in “source-sink” dynamics of Escherichia coli urovirulence. J. Mol. Evol. 2007;64:204–14. doi: 10.1007/s00239-006-0063-5. [DOI] [PubMed] [Google Scholar]
- Chen SL, Hung CS, Pinkner JS, Walker JN, Cusumano CK, Li Z, Bouckaert J, Gordon JI, Hultgren SJ. Positive selection identifies an in vivo role for FimH during urinary tract infection in addition to mannose binding. Proc. Natl. Acad. Sci. U. S. A. 2009;106:22439–44. doi: 10.1073/pnas.0902179106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Wu M, Henderson JP, Hooton TM, Hibbing ME, Hultgren SJ, Gordon JI. Genomic Diversity and Fitness of E. coli Strains Recovered from the Intestinal and Urinary Tracts of Women with Recurrent Urinary Tract Infection. Sci. Transl. Med. 2013;5:184ra60–184ra60. doi: 10.1126/scitranslmed.3005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013;5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- de Pace F, Boldrin de Paiva J, Nakazato G, Lancellotti M, Sircili MP, Guedes Stehling E, Dias da Silveira W, Sperandio V. Characterization of IcmF of the type VI secretion system in an avian pathogenic Escherichia coli (APEC) strain. Microbiology. 2011;157:2954–62. doi: 10.1099/mic.0.050005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pace F, Nakazato G, Pacheco A, de Paiva JB, Sperandio V, da Silveira WD. The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect. Immun. 2010;78:4990–8. doi: 10.1128/IAI.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U. (Patho-)Genomics of Escherichia coli. Int. J. Med. Microbiol. 2005;295:357–71. doi: 10.1016/j.ijmm.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Dongen S, Van Graph Clustering Via a discrete Uncoupling Process. SIAM J Matrix Anal A. 2008;30:121–141. [Google Scholar]
- Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, Chattopadhyay S, Sokurenko E, Neut C, Gower-Rousseau C, Colombel J-F, Bonnet R, Darfeuille-Michaud A, Barnich N. Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. 2013;9:e1003141. doi: 10.1371/journal.ppat.1003141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventré A, Elion J, Picard B, Denamur E. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology. 2001;147:1671–6. doi: 10.1099/00221287-147-6-1671. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejrnæs K, Stegger M, Reisner A, Ferry S, Monsen T, Holm SE, Lundgren B, Frimodtmøller N. Characteristics of Escherichia coli causing persistence or relapse of urinary tract infections: Phylogenetic groups, virulence factors and biofilm formation. Virulence. 2011;2:528–537. doi: 10.4161/viru.2.6.18189. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Dongen S, Van, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package version 3.2. Cladistics. 1989;5:164–166. [Google Scholar]
- Gnerre S, Maccallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, Berlin AM, Aird D, Costello M, Daza R, Williams L, Nicol R, Gnirke A, Nusbaum C, Lander ES, Jaffe DB. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1513–8. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad YH, Lipsitch M, Feldgarden M, Arachchi HM, Cerqueira GC, Fitzgerald M, Godfrey P, Haas BJ, Murphy CI, Russ C, Sykes S, Walker BJ, Wortman JR, Young S, Zeng Q, Abouelleil A, Bochicchio J, Chauvin S, Desmet T, Gujja S, McCowan C, Montmayeur A, Steelman S, Frimodt-Møller J, Petersen AM, Struve C, Krogfelt Ka, Bingen E, Weill F-X, Lander ES, Nusbaum C, Birren BW, Hung DT, Hanage WP. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc. Natl. Acad. Sci. U. S. A. 2012;109:3065–70. doi: 10.1073/pnas.1121491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. Analysis of the Genome Structure of the Nonpathogenic Probiotic Escherichia coli Strain Nissle 1917. J. Bacteriol. 2004;186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommais F, Gouriou S, Amorin C, Bui H, Rahimy MC, Picard B, Denamur E. The FimH A27V Mutation Is Pathoadaptive for Urovirulence in Escherichia coli B2 Phylogenetic Group Isolates The FimH A27V Mutation Is Pathoadaptive for Urovirulence in Escherichia coli B2 Phylogenetic Group Isolates. Infect. Immun. 2003;71:3619–3622. doi: 10.1128/IAI.71.6.3619-3622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RS, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. A type VI Secretion System of Pseudomonas aeruginosa Targets a Toxin to Bacteria. Cell Host Microbe. 2011;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Owens KL, Clabots CR, Weissman SJ, Cannon SB. Phylogenetic relationships among clonal groups of extraintestinal pathogenic Escherichia coli as assessed by multi-locus sequence analysis. Microbes Infect. 2006;8:1702–1713. doi: 10.1016/j.micinf.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Karfunkel D, Carmeli Y, Chmelnitsky I, Kotlovsky T, Navon-Venezia S. The emergence and dissemination of CTX-M-producing Escherichia coli sequence type 131 causing community-onset bacteremia in Israel. Eur. J. Clin. Microbiol. Infect. Dis. 2013;32:512–521. doi: 10.1007/s10096-012-1765-9. [DOI] [PubMed] [Google Scholar]
- Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenbach E, Bilker WB, Tolomeo P, Maslow JN. Impact of Diversity of Colonizing Strains on Strategies for Sampling Escherichia coli from Fecal Specimens. J. Clin. Microbiol. 2008;46:3094–96. doi: 10.1128/JCM.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabbett AN, Ulett GC, Watts RE, Tree JJ, Totsika M, Ong CY, Wood JM, Monaghan W, Looke DF, Nimmo GR, Svanborg C, Schembri MA. Virulence properties of asymptomatic bacteriuria Escherichia coli. Int. J. Med. Microbiol. 2009;299:53–63. doi: 10.1016/j.ijmm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Mckenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, Depristo MA. The Genome Analysis Toolkit : A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E, Johnson JR, Pérez T, Prats G, Kuskowski MA, Andreu A. Structure and urovirulence characteristics of the fecal Escherichia coli population among healthy women. Microbes Infect. 2009;11:274–80. doi: 10.1016/j.micinf.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J. Clin. Microbiol. 2008;46:2529–34. doi: 10.1128/JCM.00813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriel DG, Tan L, Goh KGK, Phan M, Ipe DS, Lo AW, Peters KM, Ulett GC, Beatson SA, Schembri MA. A Novel Protective Vaccine Antigen from the Core Escherichia coli Genome. mSphere. 2016;1:e00326–16. doi: 10.1128/mSphere.00326-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KL, Dynesen P, Larsen P, Frimodt-Møller N. Faecal Escherichia coli from patients with E. coli urinary tract infection and healthy controls who have never had a urinary tract infection. J. Med. Microbiol. 2014a;63:582–9. doi: 10.1099/jmm.0.068783-0. [DOI] [PubMed] [Google Scholar]
- Nielsen KL, Godfrey PA, Stegger M, Andersen PS, Feldgarden M, Frimodt-Møller N. Selection of unique Escherichia coli clones by random amplified polymorphic DNA (RAPD): Evaluation by whole genome sequencing. J. microbial. Methods. 2014b;103:101–103. doi: 10.1016/j.mimet.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KL, Dynesen P, Larsen P, Jakobsen L, Andersen PS, Frimodt-Møller N. The Role of Urinary Cathelicidin (LL-37) and Human β-defensin 1 (hBD-1) in Uncomplicated Escherichia coli Urinary Tract Infections. Infect. Immun. 2014;82:1572–1578. doi: 10.1128/IAI.01393-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KL, Stegger M, Godfrey PA, Feldgarden M, Andersen PS, Frimodt-Møller N. Adaptation of Escherichia coli traversing from the faecal environment to the urinary tract. J. Med. Microbiol. Int. J. Med. Microbiol. 2016;306:595–603. doi: 10.1016/j.ijmm.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norinder BS, Köves B, Yadav M, Brauner A, Svanborg C. Do Escherichia coli strains causing acute cystitis have a distinct virulence repertoire? Microb. Pathog. 2011;52:10–16. doi: 10.1016/j.micpath.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer ELL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schembri Ma, Kjaergaard K, Sokurenko EV, Klemm P. Molecular characterization of the Escherichia coli FimH adhesin. J. Infect. Dis. 2001;183(Suppl):S28–31. doi: 10.1086/318847. [DOI] [PubMed] [Google Scholar]
- Schwartz DJ, Kalas V, Pinkner JS, Chen SL, Spaulding CN, Dodson KW, Hultgren SJ. Positively selected FimH residues enhance virulence during urinary tract infection by altering FimH conformation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1–8. doi: 10.1073/pnas.1315203110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokurenko EV, Chesnokova V, Doyle RJ, Hasty DL. Diversity of the Escherichia coli Type 1 Fimbrial Lectin : DIFFERENTIAL BINDING TO MANNOSIDES AND UROEPITHELIAL CELLS. J. Biol. Chem. 1997;272:17880–17886. doi: 10.1074/jbc.272.28.17880. [DOI] [PubMed] [Google Scholar]
- Sokurenko EV, Chesnokova V, Dykhuizen DE, Ofek I, Wu XR, Krogfelt Ka, Struve C, Schembri MA, Hasty DL. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8922–6. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokurenko EV, Courtney HS, Maslow J, Siitonen A, Hasty DL. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J. Bacteriol. 1995;177:3680–3686. doi: 10.1128/jb.177.13.3680-3686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokurenko EV, Feldgarden M, Trintchina E, Weissman SJ, Avagyan S, Chattopadhyay S, Johnson JR, Dykhuizen DE. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol. Biol. Evol. 2004;21:1373–83. doi: 10.1093/molbev/msh136. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Durbin R. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–20. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Tartof SY, Solberg OD, Manges AR, Riley LW. Analysis of a Uropathogenic Escherichia coli Clonal Group by Multilocus Sequence Typing. J. Clin. Microbiol. 2005;43:5860–5864. doi: 10.1128/JCM.43.12.5860-5864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui MEl, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf C, Saint, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Rocha EPC, Denamur E. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman SJ, Beskhlebnaya V, Chesnokova V, Chattopadhyay S, Stamm WE, Hooton TM, Sokurenko EV. Differential stability and trade-off effects of pathoadaptive mutations in the Escherichia coli FimH adhesin. Infect. Immun. 2007;75:3548–55. doi: 10.1128/IAI.01963-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Scholes D, Chattopadhyay S, Sokurenko E. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl. Environ. Microbiol. 2012;78:1353–60. doi: 10.1128/AEM.06663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 2006;60:1136–51. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Tsukamoto T, Terai a, Kurazono H, Takeda Y, Yoshida O. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J. Urol. 1997;157:1127–9. [PubMed] [Google Scholar]
- Zhang L, Foxman B, Marrs C. Both Urinary and Rectal Escherichia coli Isolates Are Dominated by Strains of Phylogenetic Group B2. J. Clin. Microbiol. 2002;40:3951–3955. doi: 10.1128/JCM.40.11.3951-3955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.