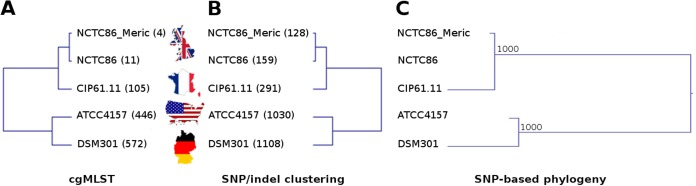
FIG 1 .
Phylogeography of ancestral Escherich strain isolates. (A) cgMLST-based UPGMA tree. The values in parentheses are numbers of allelic differences from the NCTC86_Dunne strain. (B) Hierarchical clustering (average-linkage Manhattan clustering) tree based on the presence/absence of SNPs and indels. The values in parentheses are numbers of SNP/indel differences from the NCTC86_Dunne strain. (C) Phylogenetic tree based on SNP concatenation by the maximum-likelihood method. Bootstrap values over 1,000 are indicated above the nodes. Maps of the countries of origin of the isolates filled with the respective flags are shown between panels A and B.