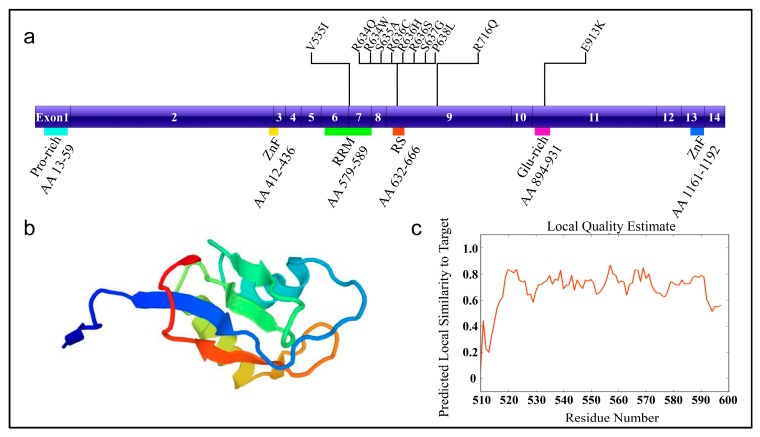
Figure 1.
The schematic diagram of RNA-binding motif 20 (RBM20) with mutation sites. (a) RBM20 structure and known mutation sites. (b) Homologous protein structure modeled with SWISS-MODEL (Q5T481, Homo sapiens) [13]. Modeled region: AA 510-597, which contains RRM domain; QMEAN = −1.56 (QMENS value indicates the degree of nativeness of the structure features and whether the model is of similar quality to experimental structures; Scores of −4.0 or below indicates very low quality); (c) The local quality plot. For each residue of the model in Figure 1b (on the x-axis), y-axis shows the expected similarity of the model to the experimental structure. The residue showing a score below 0.6 are expected to be low quality. AA: amino acid; Pro-rich: proline-rich region; ZnF: zinc finger; RRM: RNA recognition motif; RS: arginine/serine-rich domain; Glu-rich: glutamic acid-rich region; V535I: valine (V) to isoleucine (I) change at AA position 535.