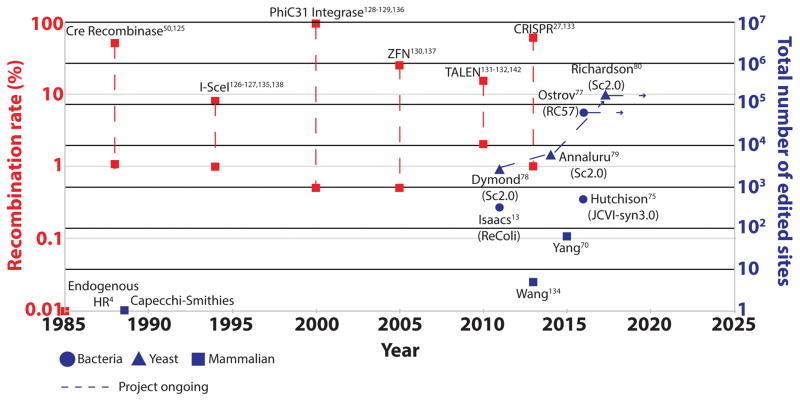
Figure 1. Timeline of gene targeting.
The timeline depicts progress in gene targeting rates through recombination (red) and the number of edited sites in a single genome (blue). Recombination rates with dashed lines denote ranges observed in the published literature4,27,28,50,58,63,64,125–147. In the mid to late 1980s, it was observed that in the presence of exogenous DNA, homologous recombination (HR) could occur at a very low frequency (0.01%). Subsequently, it was shown that by inducing a double-strand break (DSB), the rates of HR could be dramatically improved. Thus, different modalities have been discovered and employed to create DSBs at higher frequencies, from meganucleases (such as I-SceI) to zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) to the current CRISPR-associated nucleases. In addition, approaches that do not use DSB-stimulated HR such as Cre recombinase or phiC31 integrase, have also shown tremendous promise for DNA replacement, but are restricted by the limited range of sequences that can be targeted. As a corollary, in combination with the progress in DNA synthesis technologies, our improved ability to edit DNA has permitted increases in the total number of modifications being made in a single genome. In the past, 1 to 2 edits were typically made in a single genome. Ambitious projects such as RC57 and Sc2.0 have aimed to make up to four or even five orders of magnitude more edits in a single genome.