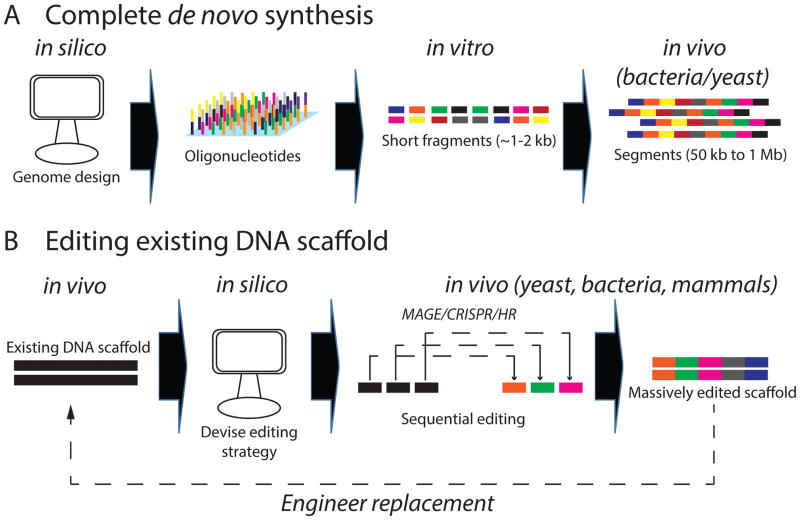
Figure 4. Two main approaches to large-scale genome engineering in bacteria and yeast.
a | For complete de novo synthesis, one must first computationally design oligonucleotide sequences that comprise the desired product while ensuring highest compatibility with the experimental strategy used for assembly. For example, thermodynamic properties of the sequence such as GC% and secondary structure formation are important parameters to consider. Subsequently, using in vitro PCR-based approaches, oligonucleotides (~150–200 bases in length) can then be assembled into fragments 1–2 kbp in size. Finally, using either bacteria or yeast as a chassis, these fragments can be assembled into segments in the order of Megabase pairs (Mbp). b | Extensive editing of an existing genome scaffold. Depending on the size of the DNA segment being edited, a segment is computationally divided into pieces with approximately similar level of editing. Each piece can then be edited independently in bacteria or yeast and upon completion, can be re-assembled into a single final segment and then placed into the final recipient cell.