Figure 6.
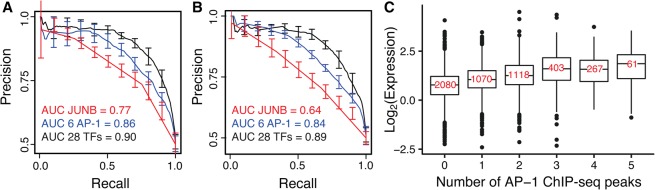
AP-1 sites that bind multiple AP-1 binding proteins in forward and reverse orientations drive high activity. (A) A logistic regression model with additive terms for motifs matches for 28 TFs can distinguish between HIGH and LOW groups of DHS AP-1-containing sequences. Precision-recall curves for models with PWMs for 28 TFs (black, AUC = 0.9), six AP-1 PWMs (JUNB, MAF::NFE2, NFE2l2, FOSL1, MAFK, NFE2; blue, AUC = 0.86), and JUNB (red, AUC = 0.77) are shown. Error bars denote standard error from fivefold cross-validation. Motif matches in both orientations were included. (B) Ignoring orientation information of motifs reduced the predictive power of logistic regression models, especially for a model with JUNB alone. Precision-recall curves for models with PWMs for 28 TFs (black, AUC = 0.89), six AP-1 PWMs (blue, AUC = 0.84), and JUNB (red, AUC = 0.64) are shown again. (C) Expression of DHS sequences containing AP-1 sites in MPRA is correlated with genomic binding of AP-1 TFs to those sequences. (x-axis) Number of peaks observed in total in five ChIP-seq experiments (JUNB, MAFF, MAFK, NFE2 and FOSL1) (The ENCODE Project Consortium 2012); (y-axis) observed log2 (RNA/DNA) counts of cis-regulatory sequences. Expression distributions for sequences with three or more ChIP-seq peaks were not significantly different from each other (Wilcoxon test, Bonferroni-corrected P > 0.05); all other distributions were significantly different from each other.