Figure 3. Two OxyS molecules can bind nusG at two different sites.
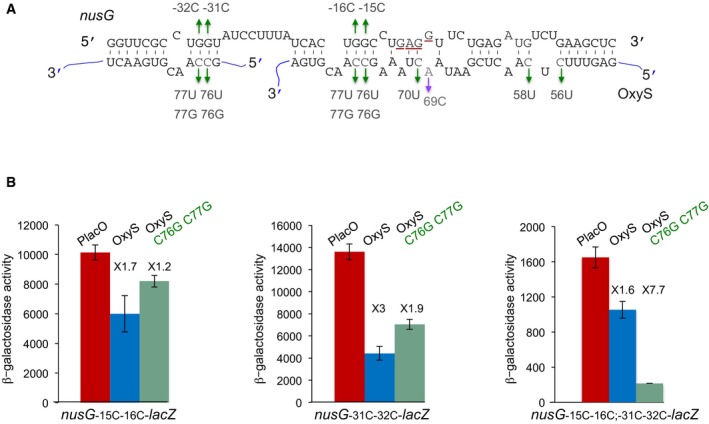
- Extended base pairing between nusG and OxyS RNAs. The initiation codon and the Shine–Dalgarno sequence of nusG are marked in red. Suppressor nontoxic OxyS mutants are in green, the highly toxic mutant is in purple. The OxyS two molecules are marked in blue. nusG quadruple mutant (G‐15C; G‐16C; G‐31C; G‐32C).
- Cultures carrying Ptac‐nusG‐lacZ (double and quadruple mutants) and OxyS (wild type and C76G; C77G) were treated with IPTG (1 mM) at OD600 = 0.1. β‐Galactosidase activity was measured 120 min after treatment. Results are displayed as mean of three biological experiments ± standard deviation. Fold repression of nusG mutants by wild type and OxyS mutant is denoted. The changes in the basal expression levels of nusG mutants could be due to an effect of the mutations on either the sequence or the structure encompassing the RBS.
