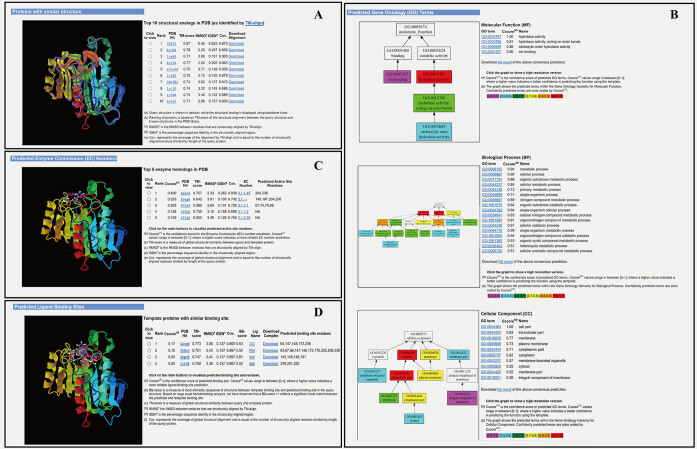
Figure 3.
An illustration of the COFACTOR web server output consisting of four annotation panels. The example is from the Escherichia coli protein ysgA (UniProt accession: P56262) with a structural model generated by I-TASSER (23). (A) Top 10 analogous structures that are structurally closest to the query structure, displaying the structural similarity between ysgA and known hydrolases. (B) GO prediction results in three aspects of molecular function (MF), biological process (BP) and cellular component (CC), which are consistent with UniProt annotation of ysgA as a putative carboxymethylene butenolidase and EcoCyc (41) annotation as a predicted hydrolase. (C) EC prediction results from top-five enzyme homologous templates, suggesting carboxymethylene butenolide hydrolase activity (EC 3.1.1.45) and directly predicting the enzyme's active site. (D) Ligand-binding site prediction results from the top 10 homologous templates, including residues surrounding putative active sites that are in proximity to the ligand. The images are screen copied from the COFACTOR example webpage (http://zhanglab.ccmb.med.umich.edu/COFACTOR/example/). Larger size copies of the images with a higher resolution are listed in Supplementary Figures S5 and S6 in the Supplementary Data.