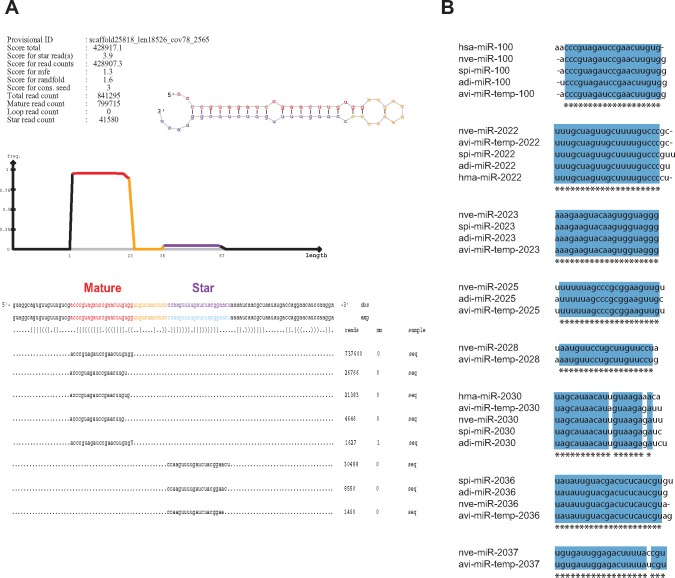
Fig. 3.
—Identified miRNAs in A.viridis with similarity to known miRNAs. (A) A typical prediction result from miRDeep2 software tool showing the miRNA precursor, mature and star sequence and their abundances in the sample. Shown is avi-miR-temp-100 precursor with top sequence alignments in the sample for each strand. (B) Alignments of novel miRNAs from A. viridis to known miRNAs from other species. Sequences of our predicted miRNAs from A. viridis (denoted as avi-miR-temp) were aligned to known miRNA sequences from H. sapiens (hsa-miR), N. vectensis (nve-miR), H. magnipapillata (hma-miR), S. pistillata (spi-miR), and A. digitifera (adi-miR). (temp = temporary; miRNAs that are not yet registered in the miRBase).