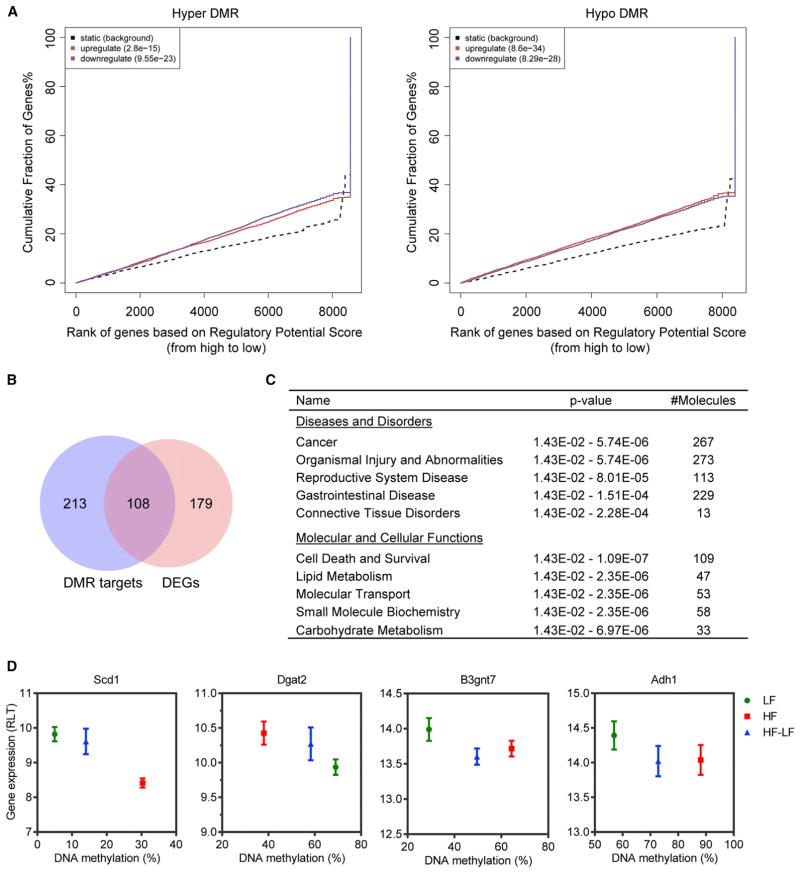
Figure 3. Obesity-Related DMRs Are Significantly Associated with DEGs.
(A) BETA with hyper-DMRs (left panel) or hypo-DMRs (right panel) and differential gene expression data (obese versus control) from young mice. The dotted line represents background genes not differentially expressed, whereas the red and the blue lines represent up- and downregulated genes in obese mice, respectively. The y axis indicates the proportion of genes in a category that are ranked at or better than the x axis value, which represents the rank on the basis of the regulatory potential score from high to low. The p values listed on the top left represent the significance of the UP or DOWN group relative to the NON group, as determined by Kolmogorov-Smirnov test.
(B) Overlap of DMR target genes with obesity-related DEGs from young mice.
(C) IPA of DMR target genes. The top five scoring hits in each functional category are shown, together with p values and the number of DMR target genes in the enriched terms.
(D) Correlation between DNA methylation and gene expression at several metabolic genes.
Error bars indicate SD. See also Figure S3.